Whether you are seeking to characterize microbial diversity in the gut, or distinguish between pathogenic and commensal bacteria on the skin, full-length 16S rRNA sequencing using PacBio systems is a valuable tool for metagenomics studies, according to microbiology researchers in a recent webinar.
PacBio microbiology expert Meredith Ashby (@AshbyMere) began the webinar by highlighting some of the strengths of the Sequel II System in metagenomics research.
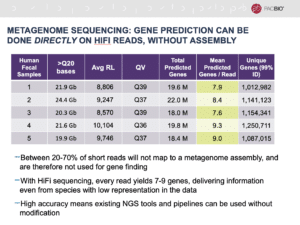
One of the biggest advantages is the ability to do gene prediction directly on high-quality HiFi reads, without the need for any assembly at all, she said. This differs from shotgun sequencing assembly using short-read data, in which anywhere between 20-70% of the data will not map to the resulting assembly.
“With HiFi sequencing, every single read yields 7-9 completely intact genes,” Ashby said. “This lets you get information even from species that have very low representation in the data, because you don’t have to have enough coverage to do assembly to improve the contiguity nor to do error correction, as is the case of other long-read technologies.”
The high accuracy of HiFi reads also means you can use existing NGS tools and pipelines, without any modification, she added.
The 16S rRNA sequencing protocol uses an asymmetric barcoded system to enable very high levels of multiplexing, and an all-in-one kit available from Shoreline Biome makes extraction, amplification and analysis even easier. Ashby said users can multiplex (at 96 or 192x) to get up to 3.6 million full-length 16S sequences. If shotgun sequencing, users can expect up to 2.4 million reads, each at about 10 kb in length.
Preemie microbiomes
Of the 450,000 pre-term babies delivered in the United States each year, about 10% born before 32 weeks develop necrotizing enterocolitis (NEC), a severe infection that happens when bacteria that are normally confined to the intestine escape through an impaired barrier, triggering a cascading inflammatory response that can lead to multi-organ failure and death in about half of afflicted babies.
In hopes of better understanding the development of the condition and the “leaky gut” that contributes to it, research associate Bing Ma, from the Institute for Genome Sciences at the University of Maryland, set out to characterize gut microbiota in preemies using 16S rRNA sequencing.
She started with short-read sequencing, and found evidence that increases in microbial diversity correlated with decreases in intestinal permeability, and higher Clostridiales abundance, in particular, was associated with an improved intestinal barrier.
“These were very exciting results,” Ma said. “However, there were limitations, and outstanding questions remained. The taxonomic resolution in these short regions of 16S was not optimal.”

To address these shortcomings in a follow-up study with an expanded cohort, Ma added full-length 16S PacBio sequencing, which picked up Bifidobacterium species that were missed by the first study. This proved important, as both Bifidobacterium and Clostriadiales were found to be linked to intestinal permeability. Ma was able to obtain species level resolution for 88% of the reads. For most of the remaining sequences, resolution was limited largely because of 16S rRNA database limitations. This high level of resolution allowed Ma to map the subspecies dynamics of B. longum and B. breve.
Ma then obtained long-read metagenomics shotgun data and was able to obtain a number of closed, circularized metagenome-assembled genomes (MAGs), including one of B. breve. The MAG of B. breve revealed a carbohydrate utilization pathway involved in the metabolism of oligosaccharides in human breast milk to short chain fatty acids. Breastfeeding and lower use of antibiotics seemed to contribute to the abundance of the bacteria, which in turn led to improved outcomes for the babies.
“Through metagenome sequencing and genome assembly, we get some mechanistic understanding of the role of the gut microbiome on the intestinal barrier maturation in early preterm neonates, and that will help us for future development of rationally formulated live biotherapeutics.”
Plenty of insights from polymorphisms
How much can you learn about the microbiome simply from polymorphisms revealed by 16S rRNA sequencing? Quite a lot, according to Jackson Laboratory PI and Human Microbiome Project leader George Weinstock (@geowei).
Weinstock first became convinced about the utility of tracking 16S polymorphisms from a 2013 study of Propionibacterium acnes. Much of the bacteria on our skin are strains of P. acnes, and Weinstock wanted to see if he could differentiate between pathogenic and commensal strains using ribotypes.
 He isolated hundreds of acne strains and conducted full-length Sanger sequencing of more than 31,000 16S rRNA gene clones. Many of the strains differed by only one or two nucleotides, but he was able to distinguish between them, and identified five that were almost exclusively found on acne lesions, and one found almost exclusively on healthy skin.
He isolated hundreds of acne strains and conducted full-length Sanger sequencing of more than 31,000 16S rRNA gene clones. Many of the strains differed by only one or two nucleotides, but he was able to distinguish between them, and identified five that were almost exclusively found on acne lesions, and one found almost exclusively on healthy skin.
“Strains can have functional differences, and sometimes the haplotypes are very tightly linked to the polymorphisms in the 16S genes, so by just looking at the 16S gene sequences, you can infer something functionally about the strains,” Weinstock said.
More recently, he used PacBio sequencing to see whether circular consensus sequencing could pick up these types of polymorphisms in E. coli, which has seven well characterized 16S rRNA genes. He sequenced a mock community of 36 strains and compared the results to both a reference sequence and a shotgun sequence of a pure E. coli isolate, with favorable results. To test clinical utility, he also compared the sequences to that of a highly pathogenic E.coli strain (E. coli 0157 Sakai).
“We could distinguish these two strains from each other even though there’s only a few mutations, looking at these polymorphisms,” Weinstock said.
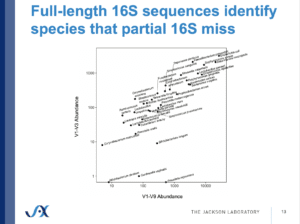
In order to confidently call such polymorphisms, however, you need high accuracy, and PacBio has become the preferred method in the Weinstock lab, he added. Similar to Ma, Weinstock has also found that full-length sequencing is able to identify species that partial 16S sequencing misses.
“The full-length sequences are definitely the gold standard, no doubt about it,” Weinstock said. “It really is worth putting the effort into full-length sequences.”
Watch the webinar:
Learn more about metagenomic sequencing with HiFi reads
July 29, 2020 | Microbiome