We’re excited to report that another team has used PacBio long-read sequencing to produce a population-specific reference genome — this time an Egyptian genome that should prove valuable for boosting precision medicine for people of North African ancestry.
Lead author Inken Wohlers, senior authors Hauke Busch (@BuschLab) and Saleh Ibrahim, and their collaborators at the University of Lübeck, Mansoura University, and other institutions report their results in Nature Communications. The need for a population-specific reference was clear: the authors note that “only 2% of individuals included in [genome-wide association studies] are of African ancestry” but “genetic disease risk may differ [between populations], especially for individuals of African ancestry.” The new assembly will serve as a foundation for more accurate interpretation of genetic risk in this population in future studies.
“With the advent of personal genomics, population-based genetics as part of an individual’s genome is indispensable for precision medicine,” the scientists note. Without a reference-grade assembly to use for comparison, people of African descent — particularly those with North African ancestry — are at risk of continued health disparities. To address this issue, the team generated a complete genome assembly of an Egyptian male based on SMRT Sequencing.
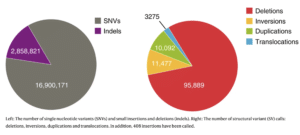
The “EGYPT” assembly spans 2.8 Gb and includes 20 Mb of novel sequence not seen in GRCh38. The authors report that the assembly is “high quality… comparable to the publicly available assembly AK1 of a Korean individual and the assembly of a Yoruba individual.”
Wohlers et al. also sequenced 110 Egyptian individuals with short reads, which were compared to the reference genome to identify population-specific variation. They called nearly 20 million single nucleotide variants and small indels, and more than 120,000 structural variants. The authors explain that it is key to understand variation as “differences in allele frequencies and linkage disequilibrium between Egyptians and Europeans may compromise the transferability of European ancestry-based genetic disease risk and polygenic scores, substantiating the need for multi-ethnic genome references,” the scientists write.
“The Egyptian genome reference will be a valuable resource for precision medicine,” the team adds. “The wealth of information it provides can be immediately utilized to study in-depth personal genomics and common Egyptian genetics and its impact on molecular phenotypes and disease.”
For more information about this project and its implications for improving genomic analysis, don’t miss this webinar and this poster from the authors.
SMRT Sequencing is being used to develop population-specific reference genomes as part of several international research efforts. Learn more about these projects and explore detailed assembly information in our interactive map.