UPDATE — September 1, 2020: This paper is now published in Genome Research.
ORIGINAL POST — April 1, 2020
In a new preprint, scientists from the National Human Genome Research Institute, the University of Washington, and other institutions describe HiCanu, a modified version of the Canu assembler designed specifically for PacBio HiFi reads. The team put the new assembler through its paces, reporting that it significantly outperformed traditional assembly methods — even getting through centromeres, segmental duplications, and other notoriously difficult regions.
As lead authors Sergey Nurk (@sergeynurk) and Brian P. Walenz, corresponding authors Sergey Koren (@sergekoren) and Adam Phillippy (@aphillippy), and collaborators report, “HiFi is a major leap forward in terms of long-read read accuracy.” They add, “As the accuracy of other long-read technologies have not exceeded 95%, the median accuracy of current HiFi reads can exceed 99.9% (>Q30), making them a promising data type for separating highly similar repeat instances and alleles.”
HiCanu applies homopolymer compression, overlap-based error correction, and tandem repeat masking to eliminate the few remaining errors in HiFi reads, resulting in 97% of reads matching perfectly to a curated reference sequence. This near-perfect accuracy helps to distinguish high-identity genomic repeats, as differences in HiFi reads can be trusted to be biological and not sequencing errors.
The new assembler generated draft assemblies of Drosophila and several human genomes. The HiCanu assemblies were all highly contiguous and extremely accurate. “On the effectively haploid CHM13 human cell line, HiCanu achieved an NG50 contig size of 77 Mbp with a per-base consensus accuracy of 99.999% (QV50), surpassing recent assemblies of high-coverage, ultra-long Oxford Nanopore reads in terms of both accuracy and continuity,” the scientists write. The reported difference in accuracy is especially large: the HiCanu assembly has 831× fewer errors than the assembly of ultra-long Oxford Nanopore reads.
The team zoomed in on certain regions known to be challenging — including centromeres, segmental duplications, and the MHC locus. For CHM13, the scientists report, “This HiCanu assembly correctly resolves 337 out of 341 validation BACs sampled from known segmental duplications and provides the first preliminary assemblies of 9 complete human centromeric regions.”
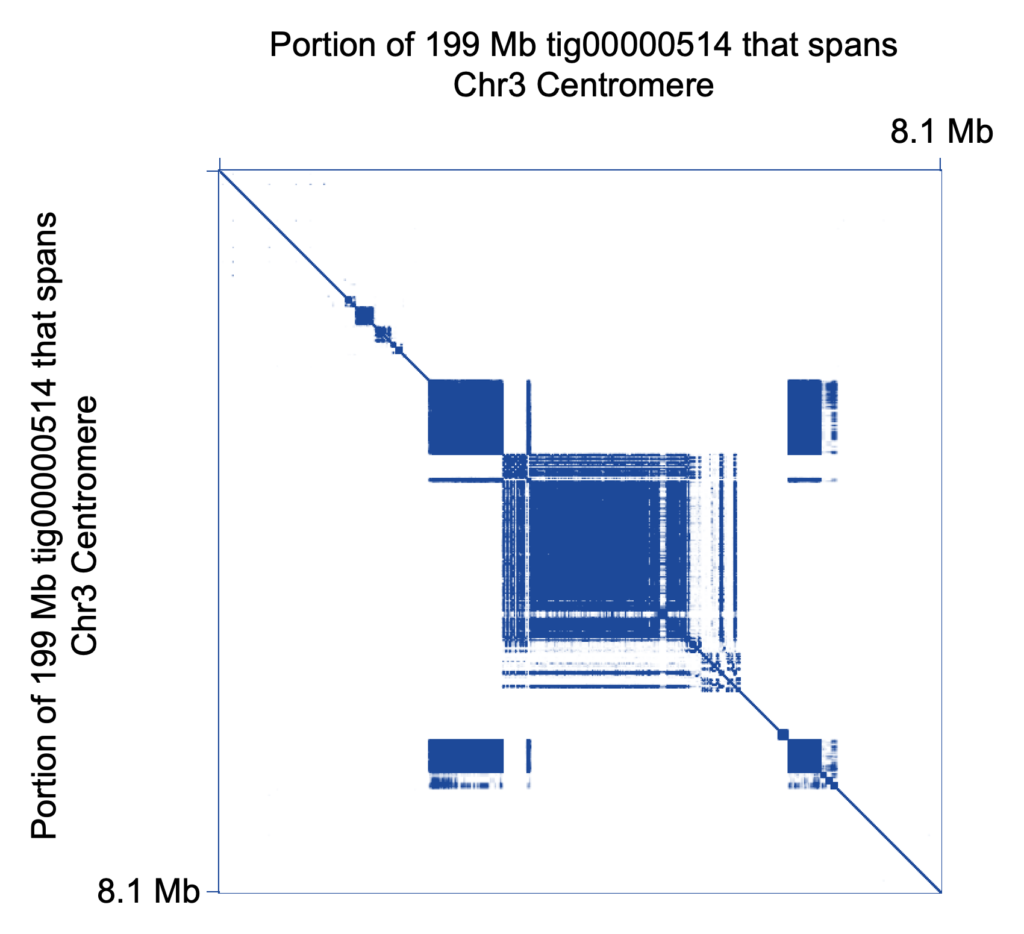
HiCanu also deftly handles haplotype phasing, with the authors stating that “HiCanu consistently recovers both haplotypes for the six canonical MHC typing genes in the human genome.”
The authors report several other advantages of HiCanu. First, assemblies generated by HiCanu do not require polishing. In fact, the authors “discourage polishing HiCanu HiFi assemblies, because… polishing pipelines may map reads back to the wrong repeat copies and actually introduce errors.” Second, HiCanu is computationally efficient: “The number of CPU hours required for assembly of a human genome is under 4,000, which could be completed on any modern cloud platform in less than a day for a few hundred dollars,” the team reports. “This is 30-fold less than recent Oxford Nanopore assemblies that required more than 100,000 CPU [hours].”
“We have demonstrated that HiCanu is capable of generating the most accurate and complete human genome assemblies to date,” the scientists write, pointing out that HiCanu could also be applied to non-human genomes, including metagenomic samples. “These results represent a significant advance towards the complete assembly of human genomes.”
Learn more about the computational approach behind HiCanu in this Medium post by Liz Tseng (@magdoll).
April 1, 2020 | Human genetics research