 When looking to understand the functional implications of genetic variability, scientists should seek out the Iso-Seq method, according to Cold Spring Harbor researchers.
When looking to understand the functional implications of genetic variability, scientists should seek out the Iso-Seq method, according to Cold Spring Harbor researchers.
In a recent paper published in Frontiers in Genetics, Doreen Ware, Bo Wang, and colleagues reviewed the state of transcript sequencing and analysis technologies, and concluded that single-molecule sequencing from PacBio provided several advantages over other methods.
A major challenge in molecular biology continues to be the complex mapping of the same genome to diverse phenotypes in different tissue types, development stages and environmental conditions, the paper states.
“A better understanding of the transcripts and expression of gene regulation is not only non-trivial but lies at the heart of this challenge,” the authors write.
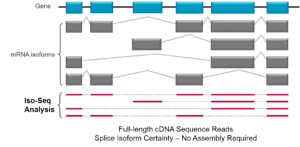
RNA sequencing can support both the discovery and quantification of transcripts using a single high-throughput sequencing assay. But methods that rely on short reads have several limitations in revealing gene regulation, the protein-coding potential of the genome and ultimately the phenotypic diversity.
Long-read SMRT Sequencing for RNA characterization has the advantage of rendering, in vitro and without ambiguity, a full-length transcript sequence without depending on the error-prone computational step of assembly. As a result, they allow a more precise detection of alternative splicing events and eventually novel isoforms, making it easier to build gene models for species which are poorly studied or have an incomplete or missing reference genome, the authors state.
“With the development of single-molecule sequencing technology, ‘one read is one transcript’ is not a dream anymore, and scientists can get the intact sequence of each isoform by sequencing a single cDNA molecule,” the authors write.
The Iso-Seq approach offers particular advantages in the characterization of polyploid transcriptomes, which have a large number of repeats and homeolog genes, and in the profiling of allele-specific expression, Ware and Wang state.
They also detail experimental and informatic pipelines and highlight several downstream applications of the Iso-Seq method, including:
- alternative splicing
- alternative polyadenylation (APA)
- fusion transcripts
- long non-coding RNAs (lncRNAs)
- isoform phasing, and
- genome annotation
Regarding the last item, the team state that the Iso-Seq method can increase the accuracy of automated genome annotation by improving genome mapping of sequencing data, correctly identifying intron-exon boundaries, directly identifying alternatively spliced transcripts, identifying transcription start and end sites, and providing precise strand orientation to single exons genes. Mapped against a reference genome, the full-length transcripts that are uncovered can be used to improve or add de novo structural and functional annotation to a genome, improve genome assembly and existing gene models, they state.
“Iso-Seq is known to retrieve longer isoforms as well as more number of isoforms… This has revolutionized our understanding of the biology of a number of organisms, including plants and animals, since transcript diversity usually represents functional diversity,” the authors write.
Iso-Seq analysis has also benefited evolutionary studies, as it allows scientists to compare the splicing variants between species and better understand the conservation of genes/isoforms, the divergence of splicing patterns, and the significance of their expression levels.
The next challenge? What to do with all the new isoforms identified from the Iso-Seq method.
The growing number of isoforms identified from different tissues/conditions within an organism will need to be ranked and prioritized for community research. And not all of them will have a meaningful impact on the cellular biological processes of the cell, Ware and Wang note, so the results will have to be carefully validated and characterized.
“Experimental approaches such as CRISPR could help by targeting the role of each isoform, and see if there are redundant or complementary functions among these different splicing isoforms,” they conclude.