Oftentimes in biology, the biggest advances come from new ways of measuring complex systems, rather than a single discovery in itself. This publication spotlight highlights a recent Nature Biotechnology paper that represents a major methodological advance in single-cell epigenomics.
In work led by researchers at the University of Washington, WashU in St. Louis, and Seattle Children’s Research Institute, the team introduces DAF-seq, short for Deaminase-Assisted single-molecule chromatin Fiber sequencing. This work demonstrates how long-read HiFi sequencing can unlock an extraordinary view of gene regulation, capturing chromatin architecture across entire chromosomes with “single-nucleotide, single-molecule, single-haplotype, and single-cell precision.”
To understand why this matters, it helps to start with the basics. In cells, DNA is packaged into chromatin, where DNA is tightly organized around structural and regulatory proteins. These proteins regulate gene activity by opening regions for activation or closing them for silencing. Gene regulation ultimately depends on where these proteins sit along the DNA and how they work together. Where this gets challenging is the hyper-variability of chromatin: It can differ from cell to cell and even between the two chromosome copies inside the same cell.
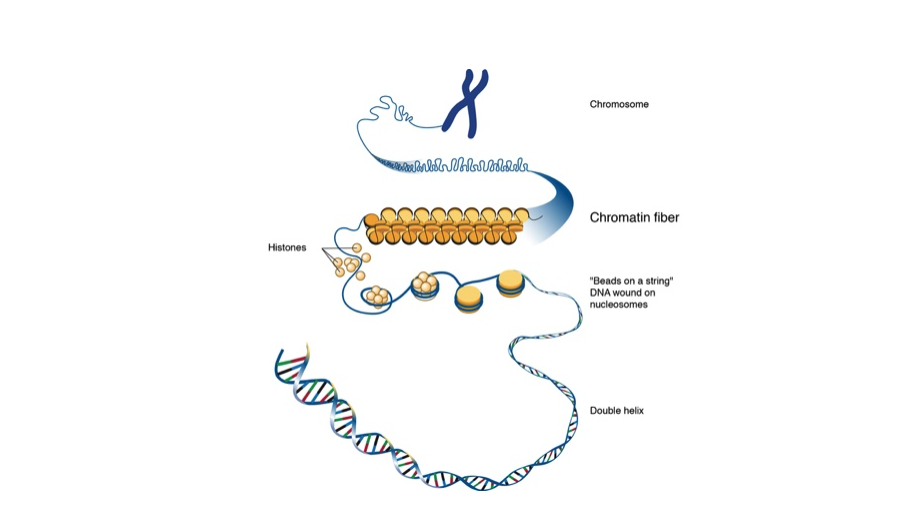
Image creator: Darryl Leja, NHGRI, creative commons
Historically, studying chromatin has required tradeoffs. Many widely used methods rely on short DNA fragments and signals averaged across large numbers of cells, which makes it difficult to know whether regulatory proteins were bound together on the same DNA molecule or simply appeared correlated when data were combined. Other approaches capture single molecules but are limited in resolution or genomic coverage. This means that we’ve had an incomplete picture of how gene regulation is organized along individual chromosomes inside single cells. The development of DAF-seq now overcomes these limitations.
DAF-seq builds on the advance of Fiber-seq, a single assay that profiles DNA sequence, methylation state, and chromatin architecture, but with a key distinction: whereas Fiber-seq avoids amplification to preserve native chromatin fibers, DAF-seq uses PCR on barcoded molecules to target regions of interest at much higher depth. This way, reads can be grouped back to their original template after PCR, avoiding amplification artifacts and retaining true single-molecule resolution. Whole genome amplification methods, like BioSkyrb Genomics’ Primary Template-directed Amplification (PTA), can be used to amplify the genome from individual cells, enabling single-cell precision.
At its core, DAF-seq is designed to reveal how proteins are arranged along individual DNA molecules. The method uses DddA, a DNA-modifying enzyme to mark accessible regions of chromatin directly on intact chromatin fibers. Regions protected by bound proteins remain unmodified, creating a detailed footprint of protein occupancy along the DNA. HiFi sequencing is central to this approach because it preserves these molecular marks while delivering long, highly accurate reads that allow chromatin state and DNA sequence to be read together on the same molecule.
The accuracy and read length of HiFi sequencing fundamentally expand what DAF-seq can achieve. On the Revio system, the researchers leverage the DddA barcoding to reconstruct the template molecule, generating ultra-long HiFi consensus reads that often exceed 100 kilobases from even single cells. By grouping reads that originate from the same original DNA strand, they reconstruct continuous chromatin fibers that can span more than 200 megabases. At this scale, a single fiber can cover most or all of a large human chromosome, with the only practical limits being chromosome length and sequencing depth. This strategy enables true single-molecule footprinting across chromosome-length chromatin fibers, representing an estimated million-fold improvement in effective resolution compared to Tn5-based methods like ATAC-Seq.
With this method, the biological findings revealed here are striking. Using DAF-seq, the authors uncover new detail about how gene regulation works at the molecular level. By directly observing which transcription factors bind cooperatively at regulatory elements at single molecules rather than averaged across many cells, this study disentangles true co-occupancy from population-level correlations and reveals precisely which proteins depend on one another and which bind independently. This method also makes it possible to directly link DNA sequence changes to their regulatory consequences. In one example, a rare somatic mutation disrupts a CTCF binding site, and DAF-seq shows how this single change leads to a loss of local chromatin organization on only the affected DNA fibers.
Perhaps most surprising is how much can be learned from very few cells. The study demonstrates how “fundamental principles of gene regulation can be derived from just 12 cells, breaking the widely accepted paradigm that high-quality single-cell chromatin analyses require data from millions of cells.” By reconstructing chromatin architectures along single fibers that can span the majority of a chromosome, the authors show that regulatory elements tend to act together over distances consistent with chromatin loops, while remaining largely independent at longer scales. These findings highlight both the plasticity and the underlying structure of the chromatin landscape.
The implications of DAF-seq extend well beyond this single study. By enabling simultaneous measurement of DNA sequence and chromatin state at single-molecule resolution, this method opens new possibilities for studying gene regulation, somatic variation, and genome organization in development and disease. Each new application built on HiFi sequencing expands its impact by enabling questions that were previously out of reach, whether through greater read length, higher accuracy, or the ability to integrate multiple layers of biological information on the same molecule.
The work showcased in this publication is another example of how our customers continue to innovate, using HiFi sequencing to achieve dramatic improvements over existing approaches and to ask questions that were previously out of reach.
Looking to go beyond traditional epigenetics approaches?
Discover how at our epigenetics page.