We are pleased to announce the launch of our new reagent kit, P6-C4, which represents the next generation of our polymerase as well as our chemistry. This kit replaces the P5-C3 chemistry and is recommended for all SMRT® Sequencing applications, including de novo assembly, targeted sequencing, isoform sequencing, minor variant detection, scaffolding, long-repeat spanning, SNP phasing, and structural variant analysis.
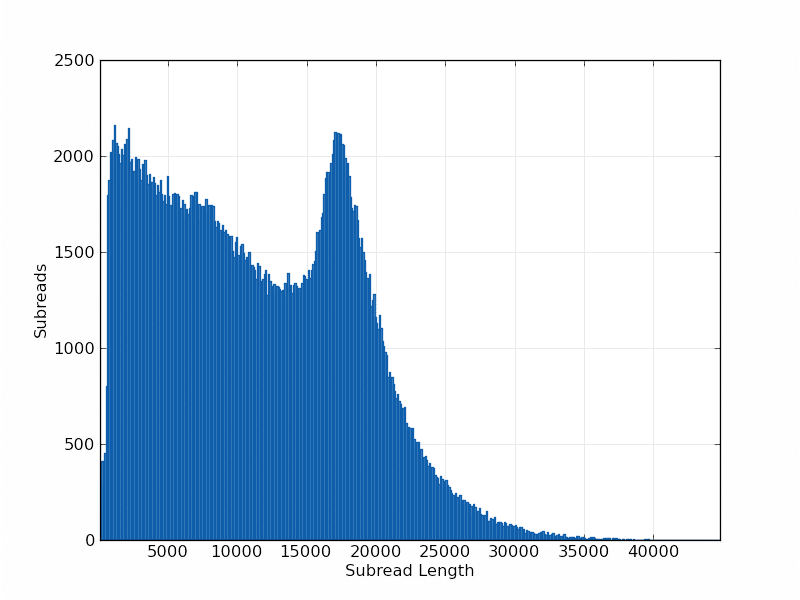
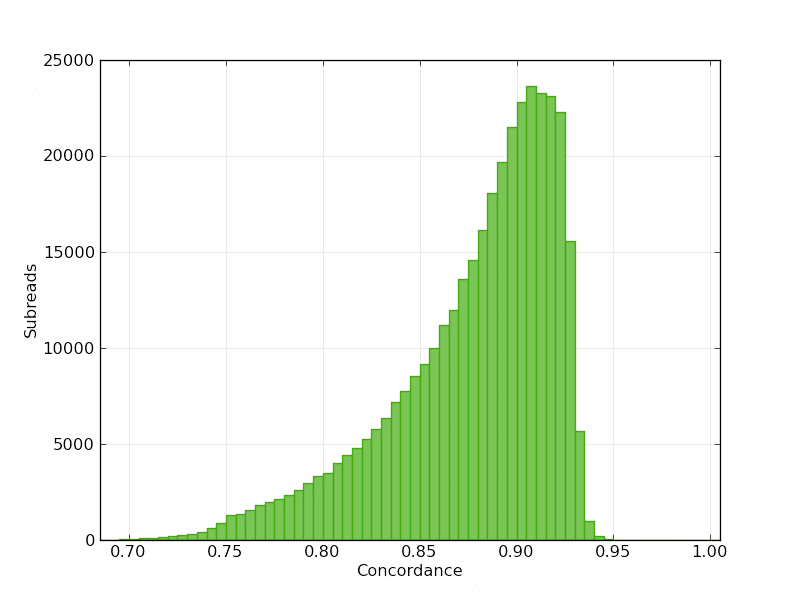
P6-C4 continues the steady read length improvement our users have seen since the instrument first launched. With this new chemistry, average read lengths increase to 10 kb – 15 kb, with half of all data in reads 14 kb or longer. The throughput is expected to be between 500 million to 1 billion bases per SMRT Cell, depending on the sample being sequenced. By providing more throughput per instrument run, the chemistry enables users to sequence larger genomes and observe previously undetected structural variants, highly repetitive regions, and distant genetic elements.
This new release also includes more robust analysis software, SMRT Analysis 2.3, providing improvements for Long Amplicon Analysis and the Iso-Seq™ method. Together with performance enhancements, these advances boost accuracy, speed up analysis, and provide more options for analyzing amplicons of mixed sizes such as full-length HLA Class I and II genes.
Here are the new part numbers:
DNA Sequencing Reagent 4.0 — P/N 100-356-200
DNA Sequencing Bundle 4.0 (10 Pack) — P/N 100-356-400
DNA/Polymerase Binding Kit P6 — P/N 100-356-300
DNA Internal Control Complex (P6) — P/N 100-356-500
We are also happy to release an additional model organism data set to the public, Caenorhabditis elegans. C. elegans sequence was first published in 1998 and has been updated and improved over the years. Our data was generated using 11 SMRT Cells with the new P6-C4 chemistry. The average read length of the raw data set is >14 kb, with half of the bases in reads > 21 kb and the maximum read length of 64,500 bases.
Some basic assembly stats:
- Genome size: 103.02 Mb
- Raw data: 4.57 Gb
- Assembly Coverage: 39.45x
- Polished Contigs: 245
- Max Contig Length: 3.17 Mb
- N50 Contig Length: 1.61 Mb
- Sum of Contig Lengths: 104.2 Mb
Mapped Subread Length Distribution:
Mapped Subread Concordance: