Assembly and binning of metagenome data are the first steps in many metagenomics analysis pipelines, and with good reason. Metagenome assembled genomes (MAGs) and circularized MAGs (CMAGs) allow recovery of complete genes and operons, thereby improving predictions of metabolic capacities. MAGs also provide information about gene synteny and enable better taxonomic profiling. However, as discussed in a recent review by Chen et. al. draft MAGs with poor completeness or high contamination can lead to incorrect conclusions.
One way to improve assembly completeness and contiguity is to use long-read sequencing. However, not all long reads are the same. Did you know that once read lengths are longer than most of the repeats in a genome or metagenome, incremental gains in raw read accuracy improve assemblies faster than higher coverage or even large gains in read length?
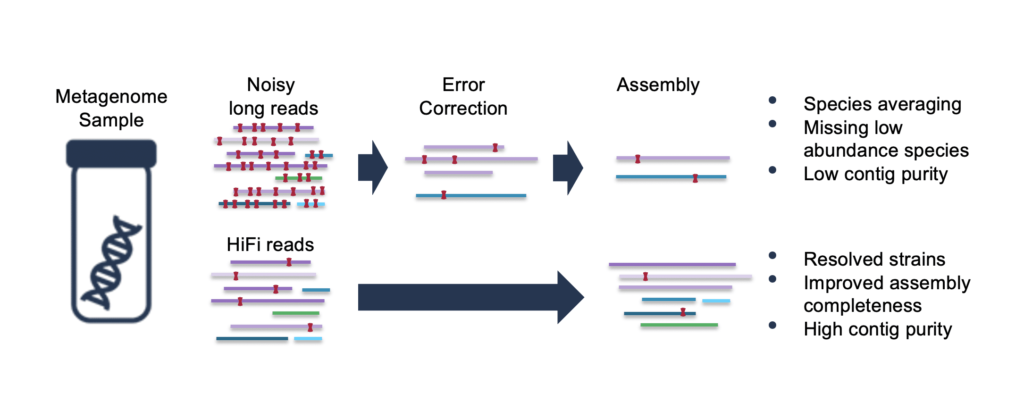
One of the main hurdles in metagenome assembly is the presence of multiple closely related strains and species in the same sample, which leads to tangled assembly graphs. While long reads are helpful in resolving these, if the difference between two bacterial species (often defined as 3%) is less than the raw error rate of your sequencing data, overlap assembly remains problematic. This is because with noisy long reads, assembly is typically preceded by an error-correction step where the raw reads are mapped against each other to produce high accuracy consensus reads.
However, with metagenome data, this has the side-effect of collapsing and averaging reads that may actually be derived from different species. The ability to distinguish reads from closely related species or strains can be effectively erased during this first step, and the purity of the resulting contigs, the completeness of the MAGs, and the total size of the metagenome assembly can all be compromised. Read on for a detailed discussion and examples of how differences in read quality impact MAG assembly.
Higher read accuracy drives assembly quality
To understand how incremental changes in accuracy and differences in coverage affect metagenome assembly quality, we generated model metagenomics datasets with community member abundances that reflect a real fecal microbiome, drawing on references from Zou, et al. and the ‘Badread’ long read simulator (Wick, 2019). Noisy long reads were simulated from 160 microbial reference genomes with accuracy modes between 87.5% and 97.5%, and HiFi reads were modeled using a typical accuracy distribution (>99%) for 8 kb -10 kb reads, an insert size commonly achievable for long read metagenome sequencing. The number of bases in each dataset was modeled after conservative Sequel II System yield of HiFi data from a metagenomics run (~20 Gb) and ONT PromethION (60 Gb) reported outputs (Shafin, 2020). The resulting model datasets were assembled with Canu 2.0, using the recommended parameters for ONT and HiFi datatypes.
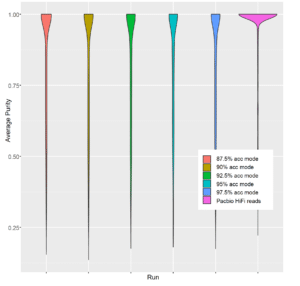
With Canu, it is possible to trace which reads were used to generate each contig in the assembly, and we used this capability to calculate the purity of each contig. Specifically, we determined what fraction of reads did not originate from the reference genome that contributed the majority of reads used to assemble that contig.
As shown in Figure 2, there are limited gains in contig purity even as accuracy changes from 85% to 97.5%. However, there is a sharp transition in contig purity when read accuracy surpasses 99%, exceeding the inter-species similarity commonly seen in a complex fecal community.
High-error reads compromise the assembly of low abundance species
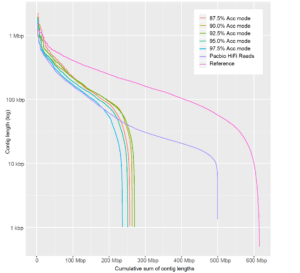 Another challenge with using self-error correction ahead of metagenome assembly relates to the uneven proportion of different species in the data. Error correction typically requires ~30-fold coverage to be effective. However, in metagenomes, it is common for species to be present at a wide range of relative abundances. This means that even when there is enough coverage of highly abundant species for error correction, reads from lower abundance species may fail the initial error correction step and be omitted from the assembly. In the example of our model data set, even with three times more raw data, the 87.5% accuracy mode dataset assembles to less than half of the expected assembly size, with contigs that are significantly shorter than with more accurate reads. When the data accuracy surpasses the threshold of microbial interspecies differences, contiguity and assembly size leap dramatically despite lower sample coverage.
Another challenge with using self-error correction ahead of metagenome assembly relates to the uneven proportion of different species in the data. Error correction typically requires ~30-fold coverage to be effective. However, in metagenomes, it is common for species to be present at a wide range of relative abundances. This means that even when there is enough coverage of highly abundant species for error correction, reads from lower abundance species may fail the initial error correction step and be omitted from the assembly. In the example of our model data set, even with three times more raw data, the 87.5% accuracy mode dataset assembles to less than half of the expected assembly size, with contigs that are significantly shorter than with more accurate reads. When the data accuracy surpasses the threshold of microbial interspecies differences, contiguity and assembly size leap dramatically despite lower sample coverage.
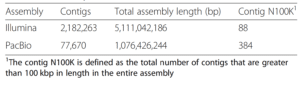 An example of how this limitation plays out in a real-world sample can be seen in a cow rumen assembly that used self-corrected PacBio CLR reads with ~89% median accuracy (Bickhart, 2019). While the PacBio CLR assembly had higher contiguity than the Illumina assembly despite a 3-fold lower depth of sequencing, the Illumina assembly had superior completeness.
An example of how this limitation plays out in a real-world sample can be seen in a cow rumen assembly that used self-corrected PacBio CLR reads with ~89% median accuracy (Bickhart, 2019). While the PacBio CLR assembly had higher contiguity than the Illumina assembly despite a 3-fold lower depth of sequencing, the Illumina assembly had superior completeness.
Closer inspection of the PacBio CLR data revealed that “the correction step removed 10% of the total reads for being singleton observations (zero overlaps with any other read) and trimmed the ends of 26% of the reads for having fewer than 2 overlaps.” The authors further noted that “this may have also impacted the assembly of low abundance or highly complex genomes in the sample by removing rare observations of DNA sequence”.
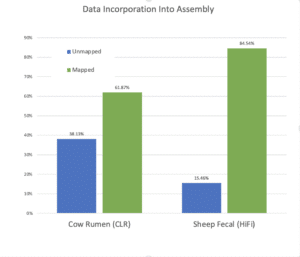
In contrast, since HiFi reads do not need error correction, all the data, including observations from low abundance species, can be used in the assembly step. Accordingly, a more recent assembly of a sheep fecal sample that used HiFi data had significantly improved performance. In his SMRT Leiden talk, Derek Bickhart noted that while cow rumen and sheep fecal samples are different communities and therefore their assemblies are not an “apple to apples” comparison, the sheep fecal assembly, done with HiFi data, appears to have a significantly improved representation of low abundance species as gauged by the proportion of same-sample short read data that maps to the long read assembly.
One possible method for overcoming the long-read coverage bottleneck is to use short read data for error correction. However, this approach suffers from the same factors that limit short read metagenome assembly. Namely, short read data has GC bias and cannot be mapped uniquely to repetitive regions. Given that bacterial genomes can range from 13-75% GC, error correcting low accuracy long reads from all the species in a metagenome sample with short read data can be problematic.
The power of HiFi reads
With the unique combination of high accuracy and long read length, HiFi data shows promise for overcoming some of the longstanding challenges in metagenome assembly. Unlike noisy long reads, assembly of HiFi reads is unencumbered by an error correction step that can erase the variation needed to correctly assemble closely related species in complex communities and generate high quality MAGs and CMAGs. Furthermore, they show potential for improving the representation and contiguity of low abundance species in metagenome assemblies.
HiFi data has already been making waves in the world of large genome assembly, first at PAGXXVIII in January 2020 and more recently at the precision FDA Truth Challenge V2, which evaluated methods for variant calling in human genomes. We are excited to see what HiFi data will do for metagenome assembly as more researchers become aware of its potential.
Learn more about HiFi sequencing for metagenomics. To start planning your metagenome assembly experiment connect with a PacBio scientist.
References:
Chen L-X, et. al. (2020) Accurate and complete genomes from metagenomes. Genome Research 30:1-19.
Bickhart, D., et. al. (2019) Assignment of virus and antimicrobial resistance genes to microbial hosts in a complex microbial community by combined long-read assembly and proximity ligation. Genome Biology 20:153.
Wick RR. (2019) Badread: simulation of error-prone long reads. Journal of Open Source Software. 4(36):1316.
Shafin, K., Pesout, T., Lorig-Roach, R. et al. (2020) Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat Biotechnol.
Zou, Y., Xue, W., Luo, G. et al. (2019) 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol 37, 179–185.
October 21, 2020 | Microbial sequencing methods