UPDATE: The paper has been published in Nature Biotechnology
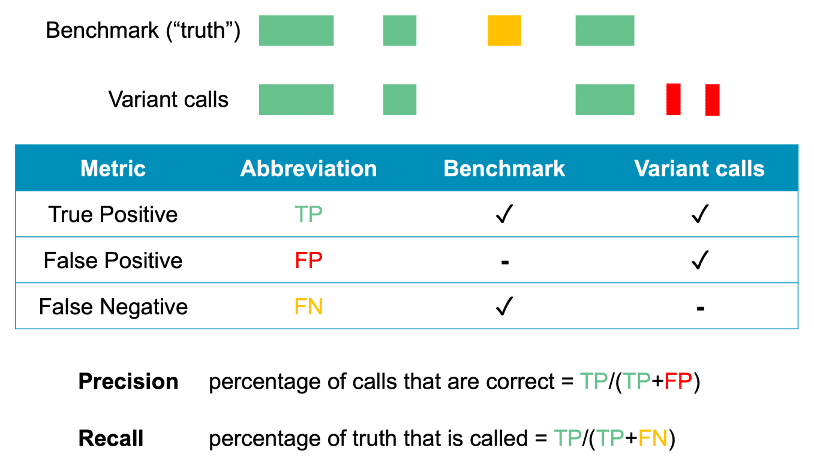
A preprint released this week from the Genome in a Bottle (GIAB) Consortium describes a benchmark set of structural variants (SVs), differences ≥50 bp, in the genome of a human male named HG002. The GIAB benchmark is the first to allow measuring precision (false positives) and recall (false negatives) of different approaches to detecting structural variants. The GIAB Consortium also developed a tool, Truvari, to support evaluation of variant call sets against the benchmark.
Earlier GIAB benchmarks, first released in 2014 and last updated in 2017, have led to enormous improvements in the quality and consistency of calling single-nucleotide and small indel variants. However, due to prior limits of DNA-sequencing technologies, the benchmark has not included SVs, which are fewer in number than small variants but in total cover more base pairs.
To extend the benchmark to SVs, the GIAB Consortium sequenced HG002 and his parents with short-, linked-, and long-read technologies; analyzed the reads with 26 different software variant callers; and integrated the different methods into a final set of 12,745 high-confidence SVs across 2.69 Gb of well-characterized “Tier 1” regions in the 3 Gb human genome. The high-confidence SVs match the expected size distribution for a human genome, with the number of variants decreasing with variant size except at the size of ALU (300 bp) and LINE1 (6 kb) repeats. The high-confidence SVs also show nearly perfect Mendelian consistency, with the genotype in HG002 being consistent with inheritance from his parents.
PacBio long reads, which provide high precision and recall for structural variants, were particularly important to the benchmark. GIAB required support from PacBio long reads for all of the high-confidence variants. Further, GIAB reports “many SVs only detectable with long reads [especially in tandem repeats]” and concludes “[t]hese results confirm the importance of long read data for comprehensive SV detection.”
If you would like to use the benchmark to evaluate how well you detect SVs, GIAB provides DNA reference material and datasets, including 32-fold coverage of accurate long reads from the PacBio Sequel II System. We also offer a tutorial on how to use the GIAB datasets and the SV benchmark to evaluate precision and recall.
In the future, the GIAB Consortium plans to extend the SV benchmark to the other genomes in its portfolio, namely HG001/NA12878 and HG005. They also plan to incorporate new data, such as highly-accurate long HiFi reads from PacBio, to improve the quality and scope of the benchmark.
June 11, 2019 | Human genetics research