In the drive for precision medicine, enabling pharmacogenomics (PGx) research to investigate how an individual’s genetic variants can impact their response to medication is critical. With robust coverage of difficult-to-sequence and difficult-to map regions, combined with high accuracy variant calling and unambiguous haplotype resolution through direct phasing, HiFi sequencing is a powerful tool in PGx.
Single genes, big impacts
While microarrays, qPCR, and short-read sequencing are often used as low-cost methods for interrogating PGx loci, these technologies are unable to fully capture and phase complex loci such as CYP2D6, which metabolizes up to 25% of known medications. With updated and new approaches for targeted sequencing, accessing complex PGx loci with PacBio is cost effective and high throughput. Three recent manuscripts highlight how researchers are turning to HiFi sequencing to fully resolve such regions.
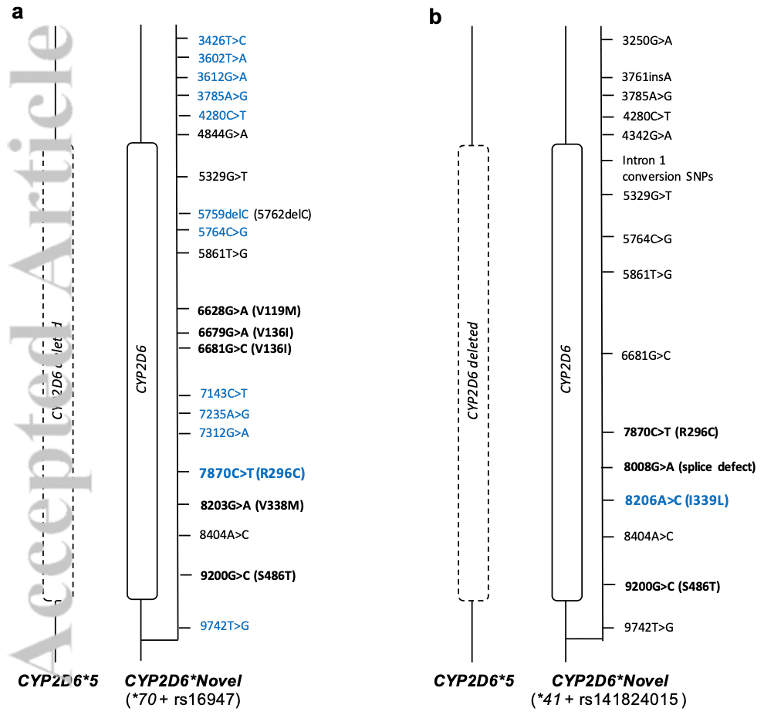
In a paper by Charnaud, et al. out of Melanie Bahlo’s lab in Melbourne, Australia, the authors use HiFi sequencing to resolve CYP2D6 in 377 Samoan Islanders, a previously uncharacterized population. “Over 20% of the population have uncharacterized or novel alleles demonstrating the diversity in CYP2D6 globally, and the relative dearth of research in people not of European ancestry.” This community is considered candidates for 8-aminoquinoline, an anti-malarial drug metabolized by CYP2D6, so assessing the rate of poor and intermediate metabolizers could have large public health impacts. Using an amplicon approach, found rare and novel variants in CYP2D6, highlighting the benefits of ancestry-agnostic HiFi sequencing in PGx research. They note that “The benefits of using long-read sequencing include being able to identify known and unknown alleles, phasing of alleles, sequencing difficult regions, and identifying large structural variants.” The authors also developed a bioinformatics analysis tool called PLASTER to accurately type CYP2D6 star (*) alleles (pharmacogenomic annotation of functional haplotypes) for screening prior to drug administration.
In another recent manuscript, Twesigomwe, et al. assessed CYP2D6 across a diverse group of sub-Saharan African populations, looking at 961 short-read whole genome samples, with a subset of 141 samples sequenced with PacBio. Many samples were derived from the Human Heredity and Health in Africa (H3Africa) Consortium and the 1000 Genomes Project. The authors identified 27 novel CYP2D6 star alleles in 5% of the participants, with HiFi reads being used to fully resolve 2 of the predicted novel alleles and identify several novel suballeles. This study represents the most comprehensive investigation of this loci in sub-Saharan Africans to date, and showed largely distinct, non-uniform allele distributions across different subpopulations, including between neighboring countries and/or ethnolinguistic groups within the same country, highlighting the significant genetic diversity across Africa. Additionally, this study showed significant differences in certain functional CYP2D6 structural variant frequency compared to previously reported estimates. Overall, their work emphasizes that important variation can be missed when using biased methods for genotyping, such as arrays or short-read sequencing, that may have gaps in coverage or rely on computational imputation for phasing.
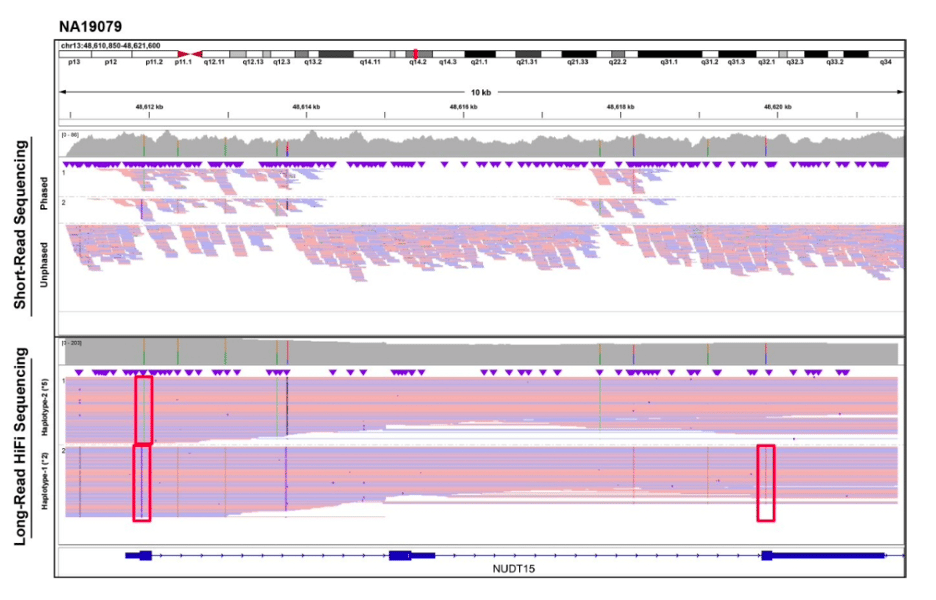
In a more clinically focused study, Scott et al. utilize HiFi amplicon sequencing for comprehensive star allele diplotyping of NUDT15. NUDT15 is involved in the metabolism of thiopurines, a class of anti-cancer and immunosuppressive therapeutics. With 3 coding exons over 8 kb, over 100 known alleles, and complex architecture, phasing is critical when interpreting variants in this gene. In a comparison of 58 Coriell samples with short-read sequencing available, only 55% of the samples could be unambiguously phased with NGS, compared to 100% with HiFi sequencing. The researchers highlight the potential clinical impact of phasing: “long-read HiFi sequencing phased all variants across the NUDT15 amplicons, including a *2/*9 diplotype that was previously characterized as *1/*2 [using other genotyping platforms].” Incorrect diplotype assignment could cause an erroneous interpretation of phenotype, which can have “dramatically different risks for thiopurine toxicity and recommended clinical management”. The HiFi assay was then used to assess the frequency of NUDT15 haplotypes in 100 Ashkenazi Jewish samples, sequenced on a single SMRT Cell, with novel alleles identified and submitted to PharmVar for star allele assignment.
In a related report, the Association for Molecular Pathology Pharmacogenetics Working Group published recommendations for TPMT and NUDT15 genotyping in laboratory tests, which also highlights the need for phasing and comprehensive variant capture for reliable phenotype assignment of pharmacogenes. This is also emphasized in similar recommendations for CYP2D6, CYP2C19, and CYP2C9 testing.
See for yourself
As seen in these recent papers, amplicon sequencing is a powerful and efficient method for capturing a single gene or small panels of interest. If you can amplify it, we can sequence it full-length with HiFi reads. With the latest SMRTbell prep kit 3.0 for amplicons protocol and Application Brief, these assays are easy to optimize and provide long-range phasing across the entire length of the amplicon, simplifying downstream data analysis. We demonstrate this workflow in a recent CYP2D6 application note, to quickly get you started with HiFi sequencing for this common PGx locus. Demo data for CYP2D6 can also be found online. For haplotyping of other PGx loci, another recent preprint highlights the development of Aldy v4, a star allele caller adapted to annotate HiFi reads for over 30 known pharmacogenes.