NOTE: This blog was updated with the latest data from the Sequel II System on March 9, 2020.
Alzheimer’s disease (AD) is a devastating neurodegenerative disease that affects ~44 million people worldwide, making it the most common form of dementia. Pathologically it is defined by severe neuronal loss, aggregation of amyloid β (Aβ) in extracellular senile plaques in the brain, and formation of intraneuronal neurofibrillary tangles consisting of hyperphosphorylated tau protein.
Studies looking into disease mechanism have shown that changes in gene expression due to alternative splicing likely contribute to the initiation and progression of AD. Hence, efforts have been made to better understand the gene expression changes in the AD brain by sequencing the transcriptome of affected brain regions.
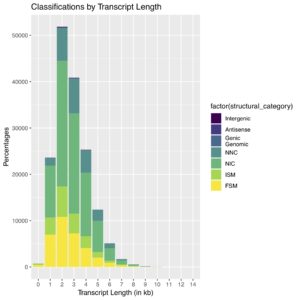
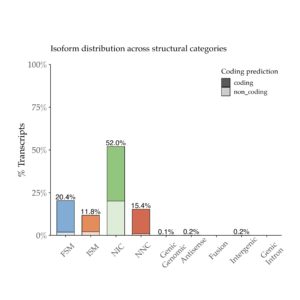
FSM = perfect match to a reference transcript. ISM = match to a reference transcript but missing one or more 5’ exons. NIC = novel isoform of known gene using known junctions. NNC = novel isoform of known gene using at least one novel splice site. For more details on transcript classification, see SQANTI2 GitHub.
Recent studies using the PacBio isoform sequencing (Iso-Seq) method demonstrated the advantages of obtaining high-quality, full-length transcript sequences for characterizing complex neural gene splicing [1], fusion cancer genes [2], and potential drug markers [3]. Here we apply the Iso-Seq method from an Alzheimer brain RNA sample. The purpose of releasing the dataset to the public is to provide researchers with a full-length transcriptome reference from which they can develop bioinformatics tools and validate their own findings.
In our final dataset, we obtained 162,290 high-quality, full-length transcript isoforms covering 17,670 genes ranging from 80 bp – 14,288 bp, with an average length of 3.3 kb (Fig. 1). When compared with the reference transcript annotation Gencode v32, almost two-thirds of the transcript isoforms are novel spliced variants (Fig. 2). Rarefaction curve analysis further showed that sampling was saturated at the gene level but novel, rare isoforms continue to be discovered (Fig. 3). The full-length transcripts revealed complex alternative splicing and fusion events (Fig. 4). A public UCSC browser session for the Alzheimer data is available.

Library Preparation and Sequencing
An Alzheimer’s Disease Brain total RNA sample was purchased from BioChain. Full-length cDNA library was generated following the Iso-Seq Express protocol. Sequencing was done on the Sequel II System using Sequel II Binding Kit 1.0 and Sequel II Sequencing Kit 1.0 using a single SMRT Cell 8M chip. Sequencing was completed in 2019.
Bioinformatics Analysis
Analysis was done using SMRT Link 8.0 “IsoSeq” protocol to generate full-length, high-quality (HQ) transcript sequences, that were then mapped to the hg38 reference genome using minimap2 (version 2.15-r905). Alignments with less than 99% coverage or 95% identity were removed and redundant transcripts were collapsed using the Cupcake tool. The SQANTI2 tool (v6.0.0) was then used to classify the collapsed, unique transcripts against Gencode v32 and filtered for potential library artifacts such as intra-priming and template switching artifacts [4].

Use and Citation
We welcome researchers to download and use the dataset for their research. For citation of the dataset, please use:
The Alzheimer brain Iso-Seq dataset was generated by Pacific Biosciences, Menlo Park, California, and additional information about the sequencing and analysis is provided at https://downloads.pacbcloud.com/public/dataset/Alzheimer2019_IsoSeq. The data used in the present study was retrieved from PacBio’s online database at https://downloads.pacbcloud.com/public/dataset/Alzheimer2019_IsoSeq (date of retrieval).
References
[1] Flaherty et al., “Neuronal impact of patient-specific aberrant NRXN1α splicing”, Nature Genetics (2019).
[2] Tian et al., “Long-read sequencing unveils IGH-DUX4 translocation into the silenced IGH allele in B-cell acute lymphoblastic leukemia”, Nature Communications (2019).
[3] Vasan et al., “Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors”, Science (2019).
[4] Tardaguila et al., “SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification”, Genome Research (2018).
September 20, 2016 | General