UPDATE: The paper is now available in Nature.
Even in the field of genomics where new breakthroughs occur every few months, completion of the first-ever fully sequenced human autosome is a momentous achievement. Highly accurate, no gaps, no mis-joins — just chromosome 8 in all its glory. It’s a remarkable feat and we are honored that PacBio HiFi reads played a pivotal role in helping to achieve it.
This work is described in a preprint recently posted to bioRxiv from lead author Glennis Logsdon (@glennis_logsdon), senior author Evan Eichler, and their collaborators in the Telomere-to-Telomere (T2T) Consortium. It is part of the broader T2T initiative to sequence and assemble the first truly complete human genome and follows the earlier release of the fully sequenced X chromosome.
“Since the announcement of the sequencing of the human genome 20 years ago, human chromosomes have remained unfinished due to large regions of highly identical repeats located within centromeres, segmental duplication, and the acrocentric short arms of chromosomes,” the authors note. “The advent of long-read sequencing technologies and associated algorithms have now made it possible to systematically assemble these regions from native DNA for the first time.”
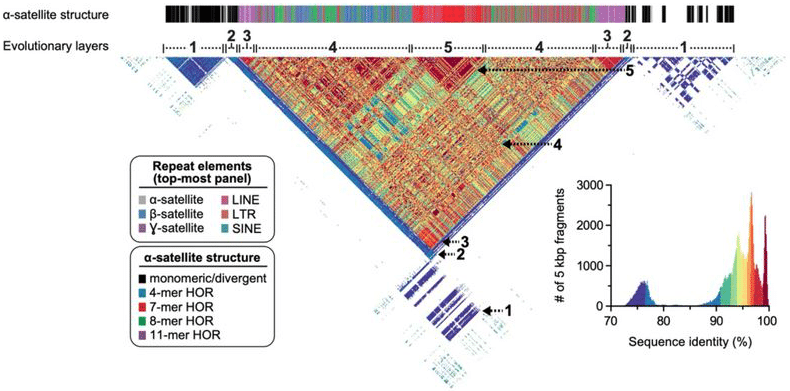
Chromosome 8 made an attractive target for the T2T’s first autosome due to its manageable centromere (previously estimated at 1.5 Mb to 2.2 Mb long). But the chromosome is also home to “one of the most structurally dynamic regions in the human genome—the β-defensin gene cluster located at 8p23.1—as well as a neocentromere located at 8q21.2, which have been largely unresolved for the last 20 years,” the scientists write. The β-defensin cluster plays a key role in innate immunity and structural variation in this region has long been implicated in human disease.
The new assembly, which addresses all five of the previously intractable gaps in the human reference genome, was built with a clever method using several data sets, including accurate long reads: “More than half of the PacBio HiFi data is contained in reads greater than 17.8 kbp, with a median accuracy exceeding 99.9%.” After a scaffolding step based on Oxford Nanopore reads, contigs assembled from PacBio HiFi reads were swapped in to provide the base-pair resolution. “We improved the base-pair accuracy of the sequence scaffolds by replacing the raw ONT sequence with several concordant PacBio HiFi contigs,” the team reports.
The complete chr8 sequence clocks in at 146 Mb and includes more than 3 Mb missing from GRCh38. As Logsdon et al. write, “The result is a whole-chromosome assembly with an estimated base-pair accuracy exceeding 99.99%.”
The scientists also tackled that persnickety β-defensin gene cluster, “which we resolved into a single 7.06 Mbp locus—substantially larger than the 4.56 Mbp region in the current human reference genome,” they note. Nearly all of that sequence data — 99.9934% of it, to be precise — came from HiFi reads. The complete centromere, meanwhile, accounted for 2.08 Mb.
With this beautiful assembly in hand, the T2T team took it out for a spin. First, they validated it with a host of orthogonal tools, such as optical mapping. Next, they generated HiFi data for the chromosome 8 orthologs in chimpanzee, macaque, and orangutan to compare the sequence data and reconstruct the evolutionary history of the human autosome. “Comparative and phylogenetic analyses show that the higher-order α-satellite structure evolved specifically in the great ape ancestor, and the centromeric region evolved with a layered symmetry,” the team writes. “We estimate that the mutation rate of centromeric satellite DNA is accelerated at least 2.2-fold, and this acceleration extends beyond the higher-order α-satellite into the flanking sequence.”
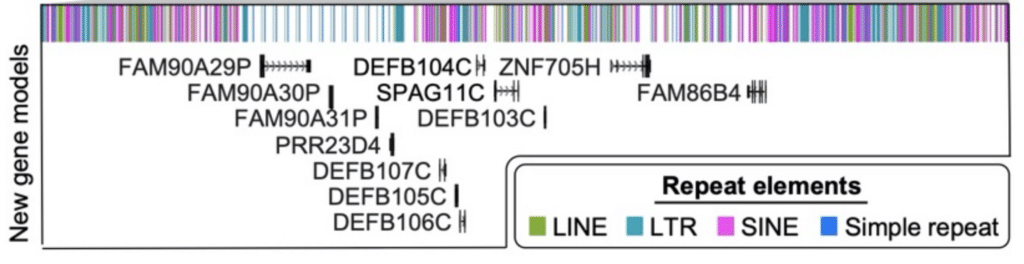
Finally, the researchers performed an analysis of full-length transcripts produced with the Iso-Seq method. That process identified “61 protein-coding and 33 noncoding loci that map better to this finished chromosome 8 sequence than to GRCh38, including the discovery of novel genes mapping to copy number polymorphic regions,” they report. Twelve of these new genes were uncovered in that tricky β-defensin locus alone.
For so many of us in the genomics community, this paper represents far more than the sequence of a single human chromosome. It’s a statement about what science can accomplish now, and where that may lead us in the years to come. As the authors summarized: “Now that complex regions such as these can be sequenced and assembled, it will be important to extend these analyses to other centromeres, multiple individuals, and additional species to understand their full impact with respect to genetic variation and evolution.”
Learn more about the research in this on-demand Labroots presentation, and find out more about whole genome sequencing and the Iso-Seq method on our site.