
Fresh off the special Nature issue last month on the first human pangenome reference (featured in a previous blog), the first regional human pangenome reference study, entitled A Pangenome Reference of 36 Chinese populations from researchers of the Chinese Pangenome Consortium (CPC), was just published in the same journal. It exemplifies a movement we have been seeing over the past few years: away from a single, linear reference to a pangenome built from many reference-quality genomes – all enabled by PacBio HiFi sequencing.
Noting that “human genomics is witnessing an ongoing paradigm shift from a single reference sequence to a pangenome form, but populations of Asian ancestry are underrepresented”, the authors present “first phase of the Chinese Pangenome Consortium, including a collection of 116 high-quality and haplotype-phased de novo assemblies based on 58 core samples representing 36 minority Chinese ethnic groups.”
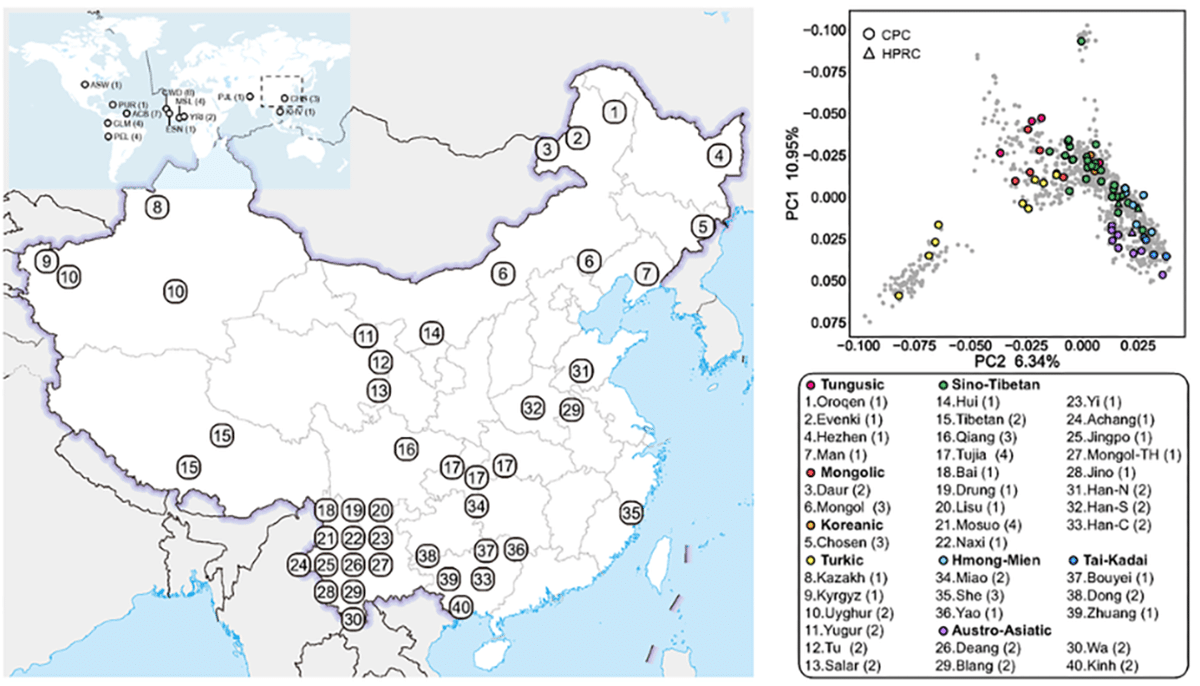
Sequencing “to an average of 30.65× using PacBio HiFi long-reads sequencing”, the authors observe that with an average contiguity N50 > 35.63 Mb and an average total size of 3.01 Gb, the “CPC assemblies largely matched or exceeded the continuity and base-level accuracy of the current reference human genome sequence (GRCh38).” Significantly, the data “add 189 million base pairs of euchromatic polymorphic sequence and 1,367 protein-coding gene duplications to GRCh38.”
Highlighting the benefits of a higher-resolution regional pangenome and “the necessity of high-quality population-specific assemblies for genetic and medical applications”, the authors identified “15.9 million small variants and 78,072 structural variants, of which 5.9 million small variants and 34,223 structural variants were not reported in a recently released pangenome reference.” Further, they “identified variations showing considerable differentiation among different ethnic groups, probably resulting from divergent ancestral backgrounds.” They also observed some genes such as CFC1, CTAG1A, and ZNF658 with highly differentiated CNVs between the CPC and HPRC assemblies.”
Stressing a “clear need to shift from a single reference to a pangenome form that better represents genomic diversity, or allelic variation, within and across human populations”, the authors conclude that “with the advancement of long-read sequencing technologies as well as computational methods, it is now feasible to enable pan-genomic construction to capture the missed variations from a large collection of diverse genomes.”
We are seeing this movement not just in human genetics, but across the tree of life – check out these plant & animal pangenome papers as just a few recent examples:
- Origin Matters: Using a Local Reference Genome Improves Measures in Population Genomics
- Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species
- Resolution of structural variation in diverse mouse genomes reveals chromatin remodeling due to transposable elements (featured in a previous blog)
Please connect with us to discuss how you can use PacBio HiFi sequencing to build a pangenome for your population or species.