
Kinnex has been making a splash since the day it launched – showing up in talks, turning heads in RNA labs, and putting its use cases solidly in the “wait, you can do that?” category of innovation. The excitement is driving adoption, and we couldn’t be more proud. But what really matters is how Kinnex performs under pressure.
A new study out of the University of Zurich and University of Virginia puts Kinnex to the test – directly against Illumina short reads, using matched samples and deep sequencing. And we’re happy to report that Kinnex not only holds its own, it sets a new bar for transcript-level quantification.
Traditional short-read RNA sequencing has been used widely for gene expression profiling, but it often falls short in accurately reconstructing full-length transcripts, especially for genes with complex splicing patterns. The Iso-Seq method – long-read RNA sequencing using PacBio HiFi technology – can accurately characterize full-length transcripts without the need for assembly, and has been used for genome annotation, novel isoform detection, allele-specific isoform expression analysis, and more. However, the relatively low depth of Iso-Seq has prevented widespread adoption of HiFi for transcript quantification – until now.
Deep long-read RNA sequencing with Kinnex
A recent preprint by Wissel et al. used high-depth PacBio Kinnex long-read RNA sequencing to analyze the differentiation process of human induced pluripotent stem cells (iPSCs) into primordial endothelial cells. The authors paired Kinnex data with sample-matched Illumina short reads, allowing for comprehensive benchmarking of the accuracy between the two platforms. A total of 11 samples over 5 timepoints were sequenced, with short reads averaging 65M and Kinnex averaging 50M reads per sample.
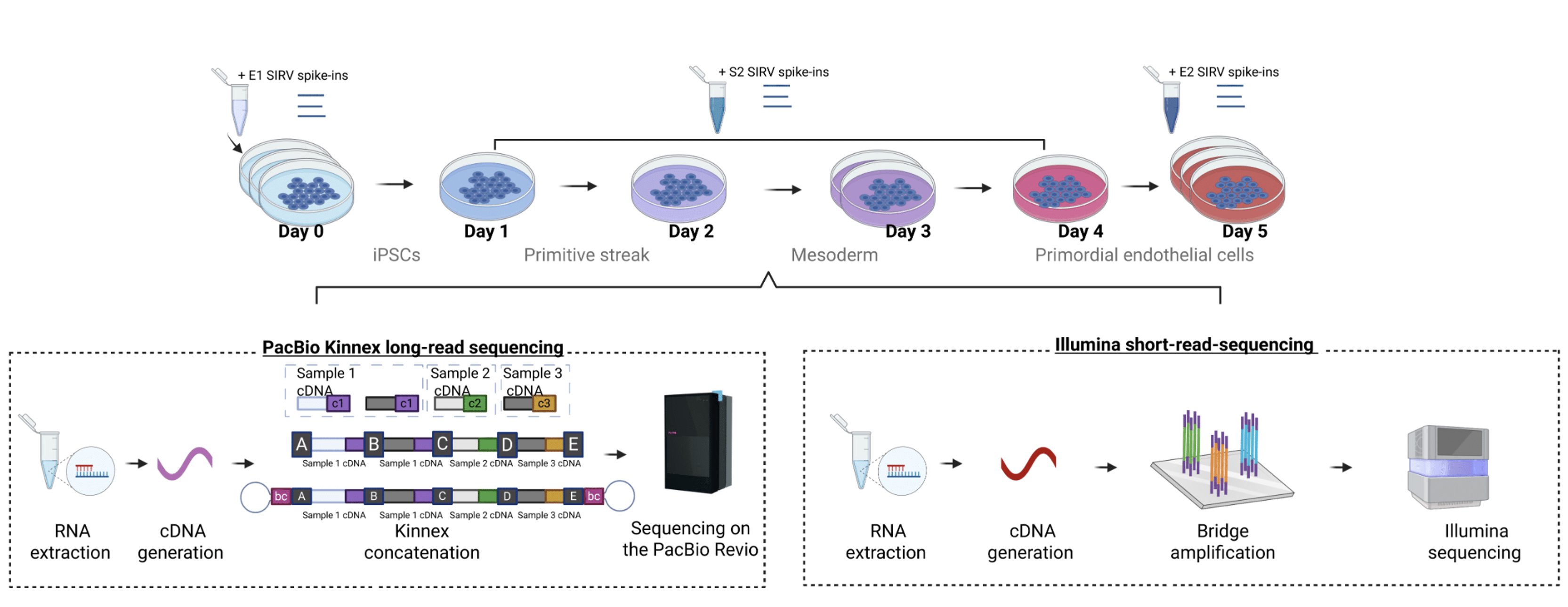
Kinnex long-read RNA sequencing data is high quality and reproducible
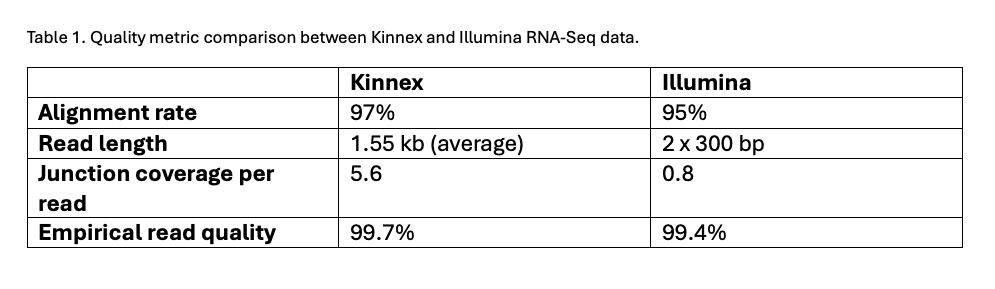
Both Kinnex and Illumina data exhibited high alignment rates, while Kinnex read lengths were longer, leading to a greater total number of bases and junctions covered (Table 1).
Both platforms exhibited overall high reproducibility of gene and transcript expressions across replicates and were able to capture the biological differentiation across timepoints.
Where short reads stumble
As the researchers discovered, the major differences between the Illumina short-read and Kinnex long-read RNA-Seq data were in accurate transcript detection and quantification in complex genes. At first glance, it appeared that Illumina detected more transcripts overall, but closer inspection revealed them to be potentially unstable or ambiguous estimates. In some cases, short read data exhibited “transcript flips”, where replicate Illumina quantifications of the same transcript fluctuated wildly, whereas Kinnex showed consistent quantification. In other cases, where short reads were unable to span multiple junctions, there is an artificial “division” of transcript quantification in short-read data, making it more difficult to determine which transcripts are actually expressed.

Because short reads struggle to resolve complex isoforms, the authors found that short reads underperformed relative to Kinnex data for detecting differential transcript expression (DTE). “The consequence of highly inferential variability in Illumina data is that it leads to inflated dispersion estimates, ultimately diminishing statistical power to detect true DTE events,” the authors wrote. When filtered for transcripts that cannot be definitively detected by short reads, both platforms showed high quantification concordance.
Kinnex long reads as a reliable choice for transcript detection and quantification
With nearly half a billion reads generated for a single WTC-11 cell line over multiple timepoints and replicates, coupled with matching deep Illumina sequencing data, this study demonstrates that Kinnex shows high reproducibility and accurate transcript detection and quantification capabilities.
Based on the results, the authors concluded that “Kinnex is a practical, reliable choice for projects that must resolve complex transcript architecture and perform routine differential expression transcript analyses.”
Curious what else belongs in the “wait, you can do that?” category?
Discover how Kinnex is redefining what’s possible in RNA sequencing – watch the webinar above or explore the product page to see it in action.