Update 1/13/14: A new data release of Arabidopsis using P5-C3 chemistry is available
Advances in our chemistries, throughput, and read length are pushing the envelope in the way we tackle larger genomes. We recently sequenced the Landsberg erecta ecotype (Ler-0) of Arabidopsis thaliana and produced a successful assembly solely using PacBio® data. The data set resulting from this sequencing effort and assembly using SMRT® Portal is now available via Devnet for anyone who wants to give it a test drive.
A few stats on Arabidopsis and the assembly using PacBio sequence data:
Genome size: 124.6 Mb
GC content: 33.92%
Raw data: 11 Gb
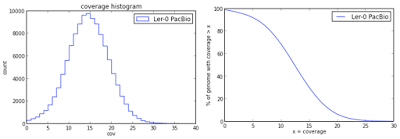
Assembly coverage: 15.37x
Polished Contigs: 540
Max Contig Length: 12.98 Mb
N50 Contig Length: 6.19 Mb
Sum of Contig Lengths: 124.57 Mb
Arabidopsis thaliana Ler-0 was sequenced using our latest P4 enzyme and C2 chemistry with a 20 Kb insert library; size selection was performed with an 8 Kb to 50 Kb elution window on a BluePippin™ device from Sage Science. We generated 11 Gb of unfiltered bases for the assembly, and used a seed read cutoff of just over 9,000 bases for preassembled reads. Assembly of the genome was performed using SMRT Portal 2.0, including polishing with Quiver. Our scientists were pleased to see that our currently available bioinformatics platform, which has demonstrated consistent utility in building high-quality assemblies for microbial genomes, worked beautifully for the more complex Arabidopsis genome as well.
We have released the input files including the preassembled reads into the Celera® Assembler for those interested in running the bioinformatics analysis and evaluation. Along with the assembled Celera results, we have also included the Quiver polished assembly for comparison. More information on the HGAP approach can be found in the recently published paper in Nature Methods.
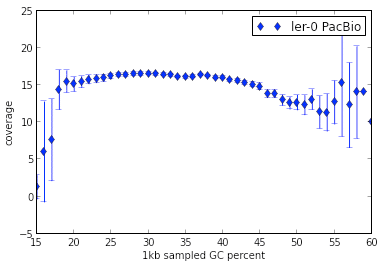
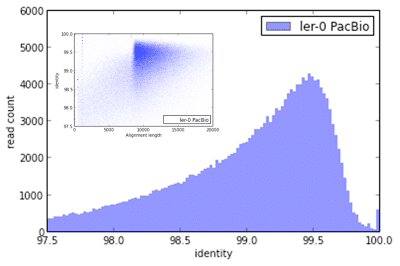
Here are some graphs summarizing the quality metrics with this genome assembly:
Distribution of sequencing coverage across assembled Arabidopsis Ler-0 genome
Coverage relative to GC content within Arabidopsis Ler-0 genome assembly
Distribution of mapped preassembled read lengths across Arabidopsis Ler-0 genome assembly
Distribution of preassembled reads mapped identity to Ler-0 genome assembly. Preassembled reads are used as input for genome assembly as part of the HGAP approach.