Scientists in China have used SMRT Sequencing to demonstrate the value of highly accurate long reads for identifying, linking, and phasing variants associated with a group of blood disorders known collectively as thalassemia. Ultimately, they predict in the Journal of Molecular Diagnostics, long-read sequencing could support a new carrier screening approach for prospective parents interested in knowing their risk of passing these diseases on to their children.
“Long molecule sequencing: a new approach for identification of clinically significant DNA variants in alpha and beta thalassemia carriers” comes from lead authors Liangpu Xu, Aiping Mao, and Hui Liu and collaborators at Fujian Provincial Maternity and Children’s Hospital, Berry Genomics, and other institutions.
The scientists undertook this project because of the need for improved carrier screening tools for ethnicities where thalassemia is prevalent, such as in Southeast Asian, Southern Chinese, Middle Eastern, Mediterranean and North African populations. While current screening methods have been fairly effective, the authors write, they “can be laborious and difficult to perform well at the laboratory level.”
Current screening techniques are unable to detect the broad range of variants and variant types associated with thalassemia. The usually fatal disease type known as α-thalassemia has been linked to nearly 130 pathogenic variants across the HBA1 and HBA2 genes, while its milder β-thalassemia counterpart can be caused by more than 200 pathogenic variants in the HBB gene. Seeking a potential approach that would provide more information about the entire spectrum of variants associated with thalassemia, the scientists turned to long-read SMRT Sequencing.
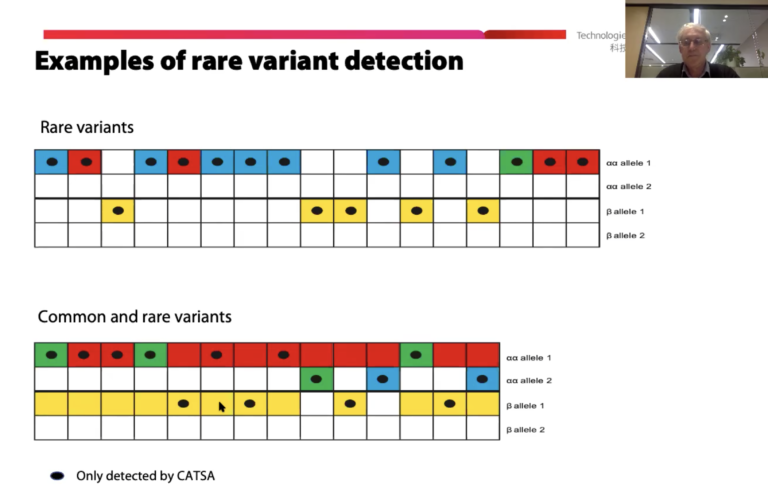
The team produced amplicons for all relevant genes. They then sequenced those regions on a Sequel System in a blinded study of 74 samples: 64 from known carriers, and 10 non-carrier controls. Results showed that “all HBA1/2 and HBB variants detected by [long-read sequencing] were concordant with those independently assigned by targeted PCR assays,” the authors report. SMRT Sequencing data correctly called the 20 known pathogenic variants in these samples. Importantly, the long-read sequencing technology “was able to discriminate compound heterozygous SNVs (trans configuration) and identify variants linked to benign SNPs (cis configuration),” the scientists add, noting that it also pinpointed linked variants which may increase accuracy of interpretation.
Based on these results, the team highlights some advantages offered by SMRT Sequencing as a possible carrier screening tool. “Since the entire gene regions are analyzed, the test has the potential to detect other HBA1/2 and HBB variants that may be outside the scope or difficult to accurately detect by traditional tests,” Xu et al. write. Also, the technology “should be capable of detecting other mild and silent HBB variants located in regulatory regions as well as HBB gene deletions that occur in approximately 1% of carriers.”
Finally, they note, being able to detect all of these variants in a single workflow with barcoded libraries makes the approach more scalable. SMRT Sequencing “displayed the hallmarks of a scalable, accurate and cost-effective genotyping methodology” that could “eventually serve as a comprehensive method for large-scale thalassemia carrier screening,” the team concludes. They also report that such an approach will become even more cost-effective when performed on the Sequel II System.
In a presentation at the PacBio Global Summit, paper co-author David Cram of Berry Genomics stated: “This type of technology would be very useful for carrier testing, not only thalassemia, but of other genetic conditions that involve complex genomic regions or copy number variants.”
December 14, 2020 | Human genetics research