Scientists have long struggled to explain the success of Mycobacterium tuberculosis in the face of effective therapeutics. Tuberculosis (TB) kills more than 1 million people annually, and has a remarkable ability to develop resistance to drugs despite its stable genome. But now, a new study from researchers at San Diego State University and other institutions strongly suggests that methylation rather than genome sequence gives M. tuberculosis its broad phenotypic range.
Lead author Samuel Modlin (@sam_modlin), senior author Faramarz Valafar (@FaramarzValafar), and collaborators report using SMRT Sequencing technology to characterize the DNA adenine methylomes of 93 clinical TB isolates. They chose samples representing diverse phylogenetic and geographic sources, and focused on methylation because of previous, smaller studies suggesting the importance of methyltransferases in M. tuberculosis. They aimed to delve deeper than those studies to see if whole methylome data could answer lingering questions about the pathogen.
“It is unclear how such a genetically static organism adapts so rapidly to drug treatment and varied immune pressures,” the scientists note. “DNA methylation is a plausible yet scarcely explored alternative mechanism for phenotypic variation in M. tuberculosis.”
In addition to producing highly accurate genetic data, SMRT Sequencing also measures epigenetic activity through kinetic changes as the DNA molecule is sequenced. The use of PacBio’s long-read technology proved critical: the long-read data enabled de novo assembly, unlike short reads that must be used with a reference-based variant calling approach. The new assemblies of the 93 isolates revealed an insertion, missed by previous studies, that is associated with an inactive methyltransferase.
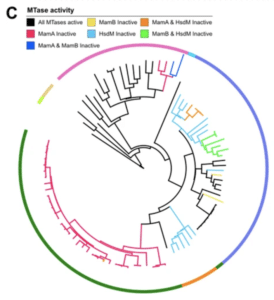
The team deployed several techniques to analyze the samples comprehensively. They produced complete, de novo genome assemblies for all isolates with SMRT Sequencing — identifying all mutations in the three methyltransferases present — and also used the kinetic data to assess methyltransferase motif sites. Phylogenetic analysis allowed them to identify epigenomic diversity across seven lineages. Finally, the team used existing transcriptomic data sets to layer onto the methylome information for a deeper analysis.
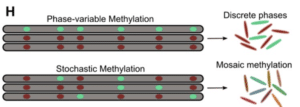
Perhaps most interestingly, the scientists used an analysis pipeline to analyze SMRT Sequencing kinetic data from each individual read, rather than in bulk. The results indicate a phenomenon the team refers to as intercellular mosaic methylation, or IMM, in which methylation is not strictly turned on or off but rather affects a subset of motif sites that vary from one cell to another.
“Mutation-driven IMM was nearly ubiquitous in the globally prominent Beijing sublineage,” they report. They also identified more than 350 hypervariable sites across the isolates where there appeared to be no consistency in methylation patterns. All told, they add, the results represent “the largest survey of methylomic diversity in [TB pathogens] to date.”
“This multi-omic integration revealed features of methylomic variability in clinical isolates and provides a rational basis for hypothesizing the functions of DNA adenine methylation in [M. tuberculosis] physiology and adaptive evolution,” the authors conclude.
“These findings add to the growing body of literature demonstrating bacterial epigenomics is an important complementary focus to genetic and phenotypic analysis in studying microbial diversity, gene regulation, and evolution.”
Learn more about methylation detection using PacBio sequencing for your research.
January 21, 2021 | Infectious disease research