UPDATE — September 3, 2021: This paper is now published in Genome Biology.
ORIGINAL POST

What does the ideal genome assembly look like? High-quality, free of errors, with no gaps, and all haplotypes resolved.
It’s a big ask, especially with challenging genomes like plants that are rich in repetitive content with high levels of heterozygosity and complex polyploidy. Moreover, such assemblies often require a combination of technologies, such as sequencing plus optical mapping.
But a team of scientists at the King Abdullah University of Science and Technology (KAUST) Core Labs (@kaust_corelabs), proved it is possible by using one technology — PacBio HiFi Sequencing — in just seven days.
Their recent preprint introduced LeafGo, a streamlined workflow able to produce a high-quality draft plant genome from plant tissue without using additional scaffolding technologies.
The rapid, one-pass approach was tested on two different Eucalyptus species, E. rudis, and E. camaldulensis.
There are more than 800 eucalypt species, but only three genomes have been published: E. grandis, E. pauciflora and E. camaldulensis. The LeafGo produced high-quality draft E. camaldulensis genome is an improvement upon those highly fragmented genomes, the KAUST team wrote.
Their assembly of E. rudis, a close relative of E. camaldulensis that inhabits a different ecological niche, is the first for that species.
“The two genomes sequenced here will improve our genomic knowledge of eucalypts, which at the moment is relatively sparse, and will assist with conservation issues and commercial uses,” they wrote.
The team tested both continuous long read (CLR) and HiFi circular consensus sequencing (CCS) data, and were especially impressed with the results from HiFi reads — “the higher base-level accuracy given by HiFi improves the assembly considerably, thus removing the need for polishing with short-read sequencing.”
“HiFi assemblies demanded less computational requirements, had higher BUSCO scores, showed several fold improvement of contig N50/N90 and L50/L90, and generated more complete genome assemblies,” the authors wrote.
“In fact, our HiFi sequencing data, assembled with hifiasm, produced near-chromosome level haploid draft genomes,” they added.
“One of the main advantages for our chosen genome assembly workflow, using hifiasm with HiFi reads, are the savings in time and compute requirements, all with minimal manual intervention.”
The estimated total time from raw reads to HiFi data to the assembly of a high-quality contiguous draft for a haploid genome of 0.6 to 1.0 Gb is approximately one day, they wrote. Assembling the HiFi data using hifiasm took 80 minutes for E. rudis (23x coverage) and 120 minutes for E. camaldulensis (27x coverage).
“When combined with time estimates of HMW DNA extraction (one day), HiFi library preparation and sequencing (five days) and assembly; a high-quality draft genome can be prepared from plant samples in seven days, depending on available compute resources,” the authors stated.
The team also created a modified Qiagen Genomic protocol in order to tackle the challenge of extracting high molecular weight DNA from the Eucalyptus species, which is difficult due to their high phenolic and polysaccharide content.
“Our extraction protocol generated high pure and copious amounts of HMW DNA within a day and using minimal resources and effort,” they wrote.
The authors say they hope LeafGo will be a valuable tool for global initiatives to sequence and assemble genomes for many thousands of eukaryotic life forms that do not yet have published standardized workflows.
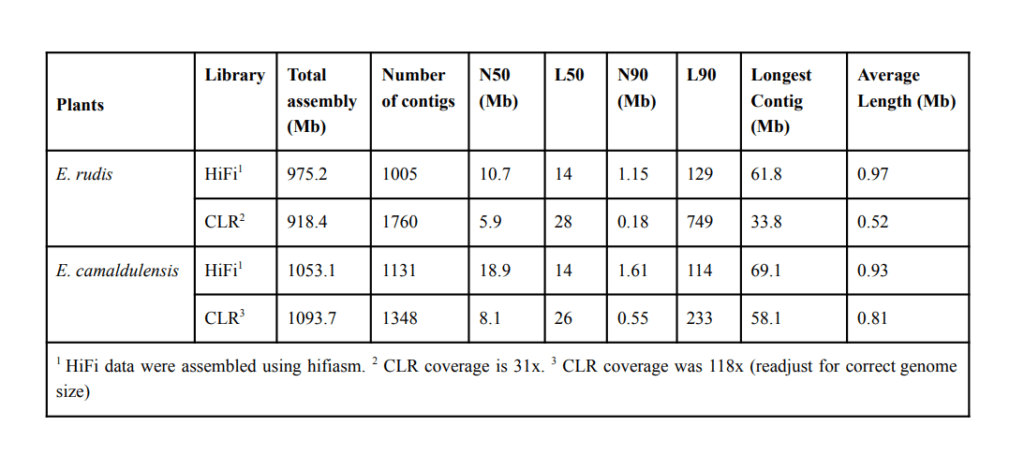
Genome assembly statistics for two Eucalyptus species