Analysis of 16S ribosomal RNA has been used for phylogenetics and identifying prokaryotes for decades. But just as scientists have had to refine the Linnaean taxonomy system based on genomic discoveries, improvements in sequencing technology are changing 16S analysis best practices.
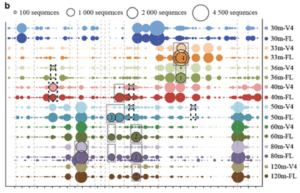
Researchers at the Joint Genome Institute, for instance, conducted a detailed benchmarking study and found that traditional methods of 16S analysis — which look at just a piece of the gene — are less accurate than analysis based on full-length sequencing of the entire V1-V9 16S gene. The full-length 16S advantage was clear in their study of the metagenome of a meromictic lake, where partial 16S sequencing led to incorrect matches or failed to differentiate the phylogenies present at different lake depths. The scientists wrote, “A resurgence of [full-length] sequences used as ‘gold standards’ has the potential to yet again transform microbial community studies, increasing the accuracy of taxonomic assignments for known and novel branches in the tree of life on previously unobtainable scales.”
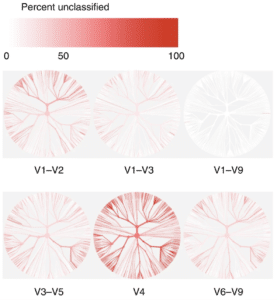
Similarly, in a recent webinar, George Weinstock (@geowei) at the Jackson Laboratory for Genomic Medicine noted that only full-length 16S sequencing can resolve all the bacterial clades commonly found in the human gut microbiome down to the species level. He said, “With V1-V9, almost all [sequences] could be accurately identified to the species level. With V4, more than half could not be identified at the species level, so you are sort of locked into the genus level or higher… The full-length sequences are definitely the gold standard, there is no question about it.”
Why does species resolution matter? Weinstock later explained using the example of 16S data from healthy stool donors. “Even though at the genus level there is a certain frequency of Bacteroides species for these samples … to do some statistical analysis based on their Bacteroides, you are going to miss a lot of important information, because the species are quite different.”
That’s where PacBio HiFi reads come in. By sequencing around and around the same molecule, HiFi sequencing produces long, highly accurate, single molecule consensus reads. At a microbiome meeting held last year at Cold Spring Harbor Laboratory, our own Meredith Ashby (@AshbyMere) presented a poster showing how HiFi reads provide both accurate and complete results for full-length 16S sequencing.
To make 16S HiFi sequencing easily accessible to PacBio users, we have developed a new and improved one-step PCR protocol and worked with several DNA sequencing service providers to add the application to their menu of services, for as little as $50 per sample. The new protocol reliably delivers more than 30,000 reads per barcode at 96-plex in a single Sequel II System run.
Here’s some information about a few of the service providers who worked with us to validate the new protocol:

DNA Services Lab, University of Illinois at Urbana-Champaign
Scientists Mark Band and Alvaro Hernandez are part of a team with extensive experience processing samples types from customers all over the world, from those that have minimal amounts of DNA, to those that have strong PCR inhibitors, such as those from corals, lakes and soils. They have 192 barcodes for generation of full-length 16S amplicons. Typical turnaround time from sample receipt to data delivery is just two weeks.

Maryland Genomics, University of Maryland School of Medicine
Part of the Institute for Genome Sciences, Maryland Genomics is led by scientists who were part of the earliest genomic efforts and who pioneered the field of metagenomics. They have expertise in working with challenging samples, particularly for microbiome studies or metagenomic applications, and operate a dedicated Microbiome Services Laboratory that provides complete sample-to-results services for full-length 16S profiling.

Biomarker Technologies, Beijing
According to CTO Liu Min, Biomarker is particularly experienced with soil and water samples for 16S sequencing, among many other types. In addition, Biomarker uses an in-house developed concatenation step to link together multiple 16S full-length amplicons before library creation, allowing them to increase throughput and reduce sequencing costs by multiplexing as many as 700 samples per SMRT Cell 8M. A current promotion offers customers 5,000 HiFi full-length 16S reads per sample for as little as $40. Learn more (Chinese language) here and here.

The University of Delaware Sequencing and Genotyping Center and Shoreline Biome, Farmington, CT
Finally, for customers who prefer an all-in-one 16S solution, Shoreline Biome offers V1-V9 and StrainID solutions that include DNA extraction, amplification, and analysis. The University of Delaware Sequencing and Genotyping Center uses Shoreline Biome technology for 16S sequencing, particularly for clinical projects such as studies of microbial communities in medical settings.
Lab director Bruce Kingham tells us, “Using PacBio long reads to resolve the 16S gene is relatively new, and has been disruptive to the field of ribotyping. The layers of genetic detail that we can elucidate from full-length 16S data could not be grasped until it was performed at the current scale, and its use continues to grow at a rapid pace.”

Mark Driscoll, the CSO of Shoreline Biome adds “Bruce’s group recognized early that the additional resolution offered by Shoreline Biome’s 2500 bp StrainID amplicon is a powerful multiplier for researchers seeking strain-level resolution beyond what is possible with the 16S gene alone. Near-perfect, contiguous HiFi reads of StrainID amplicons covering the 16S, 23S, and variable spacer between the genes enable longitudinal tracking of strains in complex fecal microbiomes in humans and model organisms such as the mouse. Researchers seeking single clone 16S sequences from their archived strain banks have been able to pack hundreds of strains in a single run.”
Ready to start planning your full-length 16S experiment? Connect with a PacBio scientist.