Whole genome sequencing
Whole genome sequencing (WGS) with PacBio HiFi sequencing delivers a more complete and accurate genome with long, single-molecule reads and 90% of bases at ≥Q30.
Contiguous and phased genome assemblies
The unique combination of length and accuracy enables HiFi reads to resolve difficult repeats, segmental duplications, and centromeres to produce more contiguous and phased assemblies.
Precision and recall for variant calling
HiFi reads also provide precisionFDA-winning accuracy for all variant types, especially in the most challenging parts of the human genome.
Methylation calling
HiFi sequencing also provides simultaneous 5mC detection in CpG contexts without any additional library preparation. This feature enables the resolution of methylation profiles with phased haplotyping and the ability to interrogate methylation abnormalities from a single sequencing experiment.
Whole genome sequencing — how PacBio compares
Human whole genome sequencing
Completely characterize human genomes to better understand the complexity of health and disease.
Plant + animal whole genome sequencing
Quickly and affordably produce contiguous, complete, and correct de novo assemblies of even the most complex genomes.
Microbial whole genome sequencing
Achieve closed chromosomes and plasmids from even the most repeat-dense and GC-rich genomes.
PacBio Compatible products
Maximize throughput by automating library prep with PacBio Compatible automation partners
PacBio Compatible Qualified automation protocols
SHAPING THE FUTURE
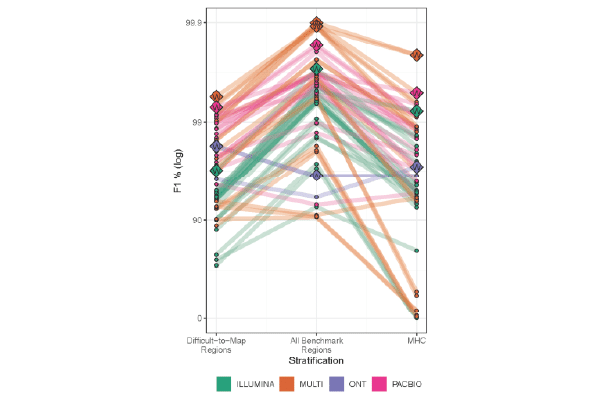
In PrecisionFDA Challenge, PacBio Hifi reads outperform both short reads and noisy long reads
PacBio HiFi reads delivered the highest precision and recall in all categories: genome-wide, specifically in difficult-to-map regions, and in the major histocompatibility complex.
Whole genome sequencing datasets
| Application | Dataset | Download literature | Technology | Sequencing system |
|---|---|---|---|---|
| Variant detection, assembly, epigenetics | Homo sapiens — GIAB trio HG002-4 | Application note | HiFi long read | Revio system |
| Tumor/normal | COLO829 melanoma | Application note | HiFi long read | Revio system |
| Tumor/normal | HCC1395 | Application note | HiFi long read | Revio system |
| Whole genome sequencing | Various plant & animals – mouse, ladybug, oak, mistletoe, and maize | Plant + animal biology | HiFi long read | Revio system |
| Assembly | Food safety & infectious microbes – 96 plex | Microbial WGS | HiFi long read | Sequel IIe system |
Whole genome sequencing applications
Variant detection
From SNVs to structural rearrangements, highly accurate single-molecule long reads allow you to confidently catalog variants of all kinds.
De novo sequencing
With HiFi reads up to 25 kb, you can efficiently and affordably generate de novo assemblies of even the most complex genomes.
Genome structure
HiFi data delivers insights into genome structure that short-read technologies often miss. Generate long reads, even through highly repetitive regions of the genome.
Epigenetics
HiFi sequencing equips you to capture epigenetic modifications during a normal sequencing run — no special chemistry or preparation required.
Workflow
PacBio library prep workflow for long-read WGS
Library preparation for WGS is a simple 5-step process with a variety of mechanical DNA shearing and size selection options
Best practice recommendations for preparing whole genome and metagenome libraries using SMRTbell prep kit 3.0 WGS
Procedure & checklist – Preparing whole genome and metagenome libraries using SMRTbell prep kit 3.0
| SMRTbell prep kit 3.0 for WGS | ||||
| Sample | Human, plant + animals | Microbial or metagenomic | ||
| DNA input per SMRT Cell per sample |
≥ 2 µg |
≥ 300 ng |
||
| DNA quality | 90% of gDNA > 10 kb | 90% of gDNA > 7 kb | ||
| SRE |
Recommended1 |
No | ||
| Mean sheared DNA size | 15 to 25 kb |
7 to 12 kb2 |
||
| Adapter indexes | SMRTbell adapter index plate 96A | SMRTbell adapter index plate 96A | ||
| Protocol automation | Yes | Yes | ||
| Manual preparation time |
4.5 hours3 |
6 hours for 24 samples4 |
||
| Size selection |
> 4 kb, bead-based |
No | ||
| Methylation data included | Yes (5mC) |
Yes (4mC and 6mA)5 |
||
|
||||
New low cost, high-throughput mechanical DNA shearing options for long-read sequencing workflows
Shearing your genomic DNA has never been easier. PacBio has developed new high throughput methods that are both cost-effective and fast to reduce bottlenecks and maximize your HiFi sequencing yields.
| Lab Equipment | Method | Throughput per run | Time per run | Mode size ranges | Consumable cost | ||||||
| Standard | Megaruptor 3 | Hydropore | 8-12 samples | 30-40 mins. | 7-25 kb | $$$ | |||||
| New | Hamilton Microlab Prep | High-speed pipette tip mixing | 8-24 samples | 22 mins. for 24 samples |
15-22 kb1 |
$ | |||||
| New | Hamilton NGS STAR/STARlet/STAR V | High-speed pipette tip mixing | 8-96 samples | 8 mins. for 96 samples with MPH |
15-22 kb1 |
$ | |||||
| New | MP Biomedical FastPrep 96 | Grinder and lysis system | 1-192 samples |
1 to 8 mins.2 |
7-20 kb3 |
$ | |||||
| New | SPEX MiniG | Grinder and lysis system | 1-96 samples |
1 to 8 mins.2 |
7-20 kb3 |
$ | |||||
|
|||||||||||
PacBio qualified automation protocols for WGS library prep and polymerase binding
PacBio has partnered with NGS automation vendors to provide scalable automated library prep methods to meet your lab’s throughput needs. From single samples on the Integra Miro Canvas, to 96 samples on the Hamilton NGS STAR or STAR V systems. Each platform protocol has been qualified through the PacBio Compatible program to ensure quality and great sequencing performance.
| Hamilton | Integra | Revvity | Tecan | |||||
| Pipette tip DNA shearing for the WGS protocol | ||||||||
| HiFi sequencing 15 to 20 kb |
NGS STAR MOA |
— | — | — | ||||
| WGS library prep protocol | ||||||||
| SMRTbell prep kit 3.0 | Miro Canvas | SciClone NGSx | DreamPrep Compact | |||||
| HiFi prep kit 96 | — | — | — | |||||
| HiFi plex prep kit 96 | — | — | — | |||||
| Anneal, binding, and clean (ABC) protocol for polymerase binding | ||||||||
| Seque II binding kit 3.2 | — | — | DreamPrep Compact | |||||
| Revio polymerase kit | — | — | DreamPrep Compact | |||||
| Revio polymerase kit 96 | — | — | — | |||||
New automation protocols will be added as they become available via our PacBio Compatible partners. Please use our automation workflow selector tool to determine which of our PacBio Compatible automation solutions are best for your application.
Whole genome sequencing in action
Case Study
Pioneering a pangenome reference collection
Learn about DuPont Pioneer’s ambitious plant pan-genome reference project.


Scaling human whole genome sequencing for rare and inherited disease research
Find out how PacBio HiFi whole genome sequencing is being used in rare disease research.
Bacterial genomes from the lakes of Minnesota to NYC hospitals
See how scientists are studying the ecophysiology of Minnesota microbes using long-read…

Uncovering structural variants associated with rare disease
Discover how Alexander Hoischen’s research group is using long-read sequencing to find…
EXPLORE
Did you know we have a comprehensive library of articles, reports, papers, and videos related to whole genome sequencing?
Whole genome sequencing workflow at a glance
Easy-to-use, high throughput sequencing for the accuracy you need at an affordable cost
Straightforward library prep
Go from sample to sequencer in a single afternoon
Complete + accurate sequencing
Obtain accurate, unbiased, and long reads
Directly detect epigenetic modifications
Universal file types
Analyze data using any of the common genome assembly and analysis tools
Common questions about PacBio whole genome sequencing
SUGGESTED WHOLE GENOME SEQUENCING PRODUCTS
Nanobind kits for DNA extraction
SMRTbell prep kit 3.0 for library prep