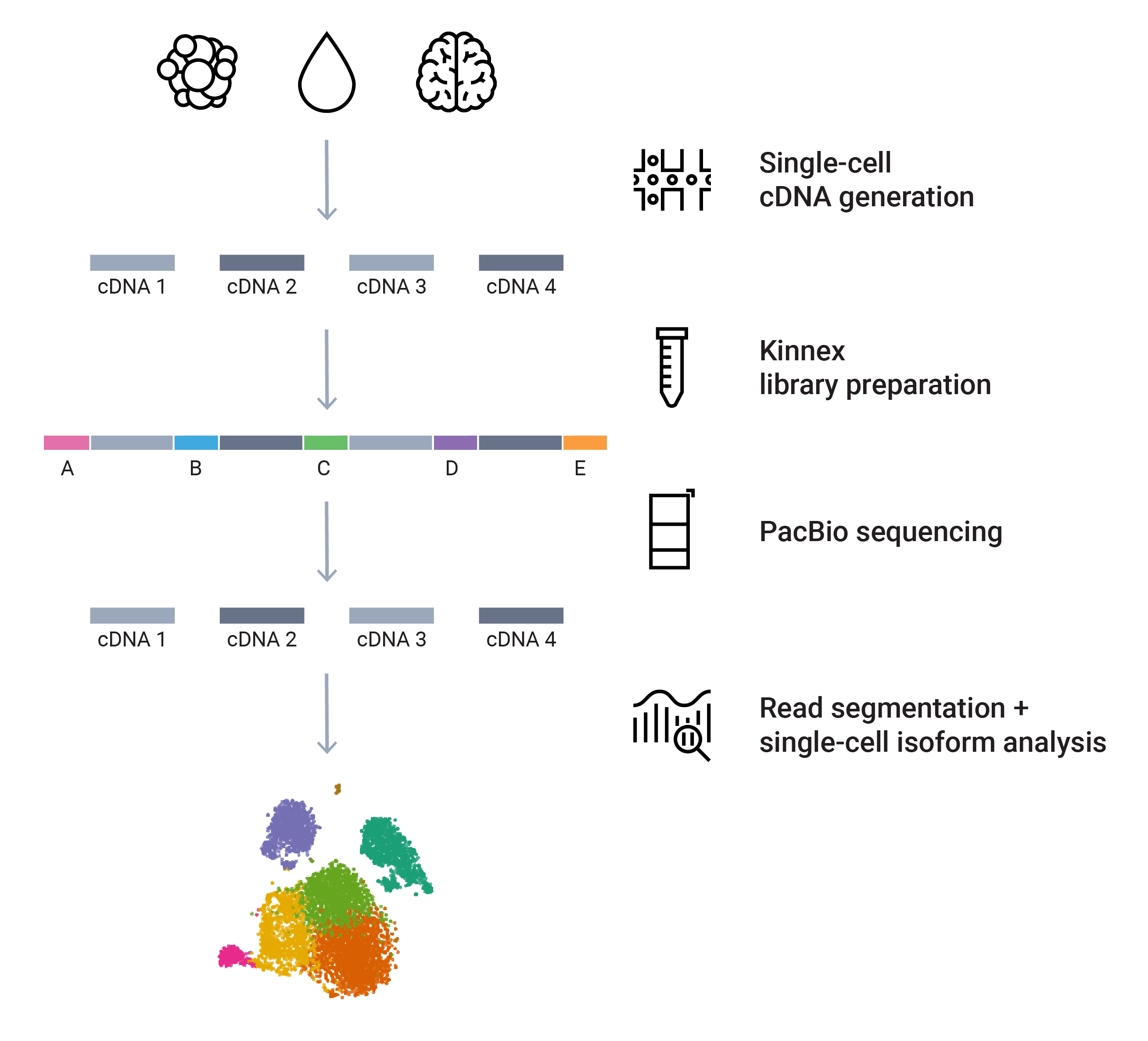
Reveal hidden isoform diversity at single-cell resolution
Understanding cell heterogeneity at the isoform level is critical for basic and disease research. Short reads can only capture gene-level information, while other long-read technologies lack the accuracy for accurate UMI/barcode identification.
HiFi reads can provide accurate detection of isoforms in your single-cell study. No complicated algorithms for barcode correction or orthogonal sequencing data are required.
With the Kinnex single-cell RNA kit, you can:
- Achieve 16-fold throughput increase on Sequel II/IIe and Revio systems
- Move from gene counting to full-length isoform information
- Reveal cell type-specific spliced isoforms and expressed variants
The Kinnex single-cell RNA kit is an upgrade of the MAS-Seq for 10x Single Cell 3’ kit that is compatible with both 10x Chromium 5’ and 3’ Single Cell cDNA and can multiplex up to four 10x libraries per run.

Video Webinar
LEARN THE MAS-SEQ TECHNOLOGY BEHIND THE KINNEX SINGLE-CELL RNA KIT: WHEN GENES ARE NOT ENOUGH
PacBio scientists present the MAS-Seq method behind the Kinnex single-cell RNA kit. Hear why isoforms, not genes, are critical for single-cell RNA research. See how highly accurate, full-length isoform information can deliver groundbreaking insights into disease biology. Finally, learn how to easily integrate Kinnex kits with a library preparation walk-through and user-friendly informatics that can be combined with downstream tools like Kana, Seurat, and CellTypist.
Tech note
DETECT RARE ISOFORMS AND CELL TYPES WITH JUMPCODE
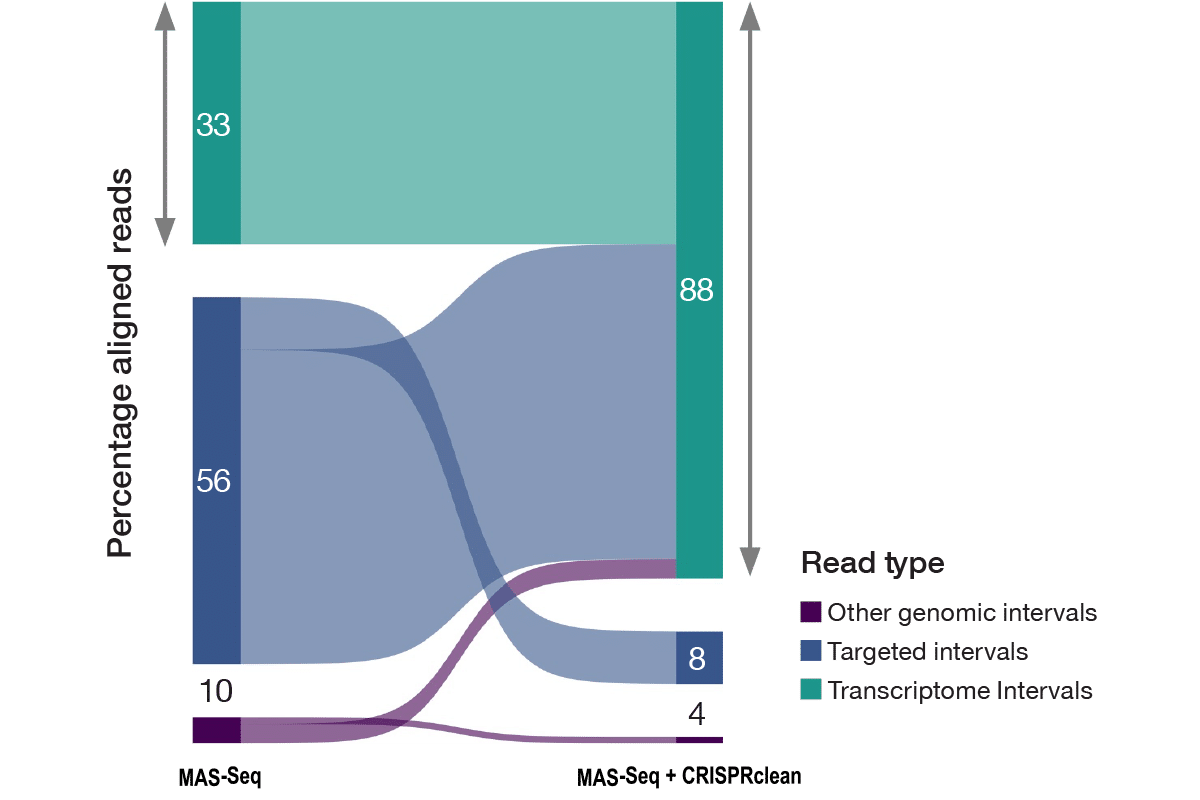
The Jumpcode Single Cell RNA Boost kit is compatible with the Kinnex single-cell RNA kit from PacBio. Using CRISPR-Cas9 technology targeting uninformative genes that are ignored by single-cell analysis, boosting discovery power of rare isoforms and cell types that otherwise cannot be seen.
|
Long-read sequencing performance |
Short reads | Other long reads | PacBio |
|---|---|---|---|
| Single technology | ✓ | X | ✓ |
| Isoform information | X | ✓ | ✓ |
| Variant information | X | ✓ | ✓ |
| Official software | ✓ | X | ✓ |
PacBio Compatible products
The Kinnex kit works with PacBio Compatible products from 10x Genomics and Jumpcode Genomics
COMMON QUESTIONS ABOUT SINGLE-CELL RNA SEQUENCING AND THE KINNEX KIT
Single-cell RNA sequencing (scRNA-seq) emerged to characterize gene expression differences between individual cells derived from a complex tissue, allowing a higher-resolution look at the transcriptome. A typical single-cell library takes RNA as input and outputs cDNA molecules that have been tagged with single-cell barcodes and unique molecular indices (UMIs) so that transcripts can be traced back to their “single cell” origins. This is in contrast with bulk RNA-Seq, where there are no single-cell barcodes to attribute transcripts back to cells.
Single-cell RNA sequencing using PacBio HiFi reads offers full-length isoform information at the single-cell level. Short-read sequencing only sequences a short fragment of one end of the transcript, providing only gene-level information. In contrast, PacBio HiFi reads can cover the entire transcript, revealing isoform diversity at the single-cell level. Isoforms — not genes — are often the driver of disease and biology.
The Kinnex single-cell RNA kit is based on the MAS-Seq method developed by Al’Khafaji et al as a concatenation method for increasing throughput by joining cDNA molecules generated from single-cell platforms into longer concatenated fragments. HiFi reads generated from sequencing the concatenated molecules can then be bioinformatically broken up to retrieve the original molecules. The result is higher throughput and reduced sequencing needs. The authors showed that, applying the MAS-Seq method to 10x single-cell libraries, unique isoforms associated with specific cell types relevant to cancer can be distinguished which are otherwise undetectable using short-read single-cell RNA-Seq.
The Kinnex single-cell RNA kit is an upgrade to the MAS-Seq for 10x Single Cell 3’ kit. It additionally supports 10x Single Cell 5’ kit (v2) and multiplexing through use of barcoded Kinnex adapters.
Yes! With MAS-Seq, you can obtain full-length isoform information for your single-cell libraries, whereas with other sequencing methods, such as short reads, you can only obtain gene-level information. Multiple publications, using MAS-Seq (Al’Khafaji et al.) or regular HiFi sequencing (Palmer et al., Joglekar et al.) have demonstrated the use of PacBio for identifying cell-type isoform specificity.
No. With the Kinnex single-cell RNA kit, PacBio HiFi sequencing offers sufficient coverage for a typical single-cell experiment with one SMRT Cell on the PacBio long-read systems. No orthogonal sequencing technology is required.
The Kinnex single-cell RNA kit is compatible with cDNA generated using the 10x Chromim Next GEM Single Cell 3’ kit (v3.1) and 5’ kit (v2).
The Kinnex single-cell RNA kit is compatible with Sequel II/IIe and Revio systems.
The SMRT Link Read Segmentation & Single-cell Iso-Seq workflow processed Kinnex single-cell data to produce gene- and isoform-count matrix that is compatible with common tertiary analysis tools such as Seurat. Further bioinformatics information can be found at https://isoseq.how/
Single-cell transcriptomics for cancer research
A more complete transcriptome for cancer research – now at single-cell level. With Kinnex, upgrade your single-cell cancer research to:
- Discover isoforms as potential biomarkers and help enable therapeutic targets
- Identify fusion isoforms
- Detect expression mutations
Workflow
EMPOWER YOUR SINGLE-CELL TRANSCRIPTOME STUDY WITH KINNEX SINGLE-CELL RNA KIT
- 80–100 million reads per Revio SMRT Cell to characterize cell type isoform expressions
- Compatible with Sequel II/IIe and Revio systems
- SMRT Link workflow outputs gene- and isoform- matrix compatible with tertiary software