Targeted sequencing
Targeted sequencing with HiFi reads allows you to sequence only the genomic regions you care about — easily and cost-effectively at scale. Hybrid capture, amplicon, and PCR-free PureTarget methods are available to suit your needs, giving you all the benefits you expect from HiFi reads including accurate haplotype resolution and comprehensive detection of all variant types. The length and accuracy of HiFi reads allow you to access genomic dark regions such as areas of high homology, GC-rich and repetitive regions, and to resolve complex gene families like HLA and pharmacogenes like CYP2D6, and sequence full-length repeat expansion targets with methylation.
- Cost-effective and flexible scaling
- Robust coverage of difficult-to-sequence or difficult-to-map regions
- Ancestry-agnostic discovery of rare or novel variants
- High-accuracy variant calling
- Unambiguous haplotype resolution through direct phasing
- Custom panels, flexible scaling, cost-effective projects
- Consolidate legacy molecular workflows into one assay
Shine a light on dark regions of the genome with the PureTarget repeat expansion panel
Scalable solution for deep sequencing of repeat expansions
Multiplex up to 48 samples on the Revio system and 24 samples on the Sequel systems for a panel of 20 repeat expansion targets. With streamlined library prep and analysis support in SMRT Link 13.1, go from samples to answer in 3 days.
What does comprehensive genotyping look like?
- Accurate sizing of both alleles, including large repeat expansions
- Single base resolution of repeat sequence
- Simultaneous detection of methylation
- Deep coverage so you can profile mosaicism
What genes are included in the panel?
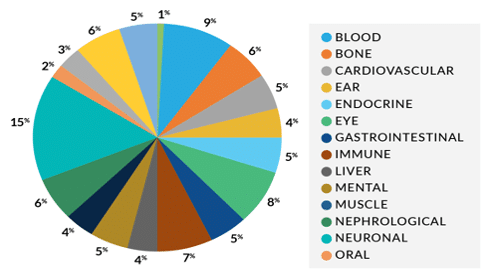
AR, ATN1, ATXN1, ATXN10, ATXN2, ATXN3, ATXN7, ATXN8, C9ORF72, CACNA1A, CNBP, DMPK, FMR1, FXN, HTT, PABPN1, PPP2R2B, RFC1, TBP, TCF4
Human genomics
Cost-effectively explore the full variation of the regions of the genome that matter to you.
HLA SEQUENCING
Overcome gene-family complexity and extreme polymorphism to identify true HLA allelic diversity.
PLANT + ANIMAL SCIENCES
Gain a comprehensive view of specific genomic regions of interest, regardless of genome size or complexity.
MICROBIAL + INFECTIOUS DISEASE RESEARCH
Resolve viral populations to better understand evolution, quasispecies dynamics, drug resistance, and immune escape
Pick the right workflow for your lab
PureTarget repeat expansion panel
A reimagined product that combines CRISPR-Cas9 technology for target enrichment and long and accurate HiFi reads to sequence the most challenging genomic regions at scale. Multiplex samples on Revio and Sequel II systems for a panel of 20 pathogenic repeat expansion targets linked to human disease. PureTarget libraries consist of DNA in its native form, including epigenetic marks like methylation and are free from errors and artifacts that can be introduced by PCR or hybridization capture methods.
HiFi Target Enrichment
Combine Twist Bioscience target enrichment with PacBio long and accurate HiFi reads to efficiently sequence your priority genomic regions at scale. Sequence enriched regions with a protocol optimized for HiFi reads to get comprehensive detection of single nucleotide variants, structural variants, and indels with haplotype resolution. HiFi target enrichment can deliver accurate alleles for complex gene families such as immune genes (e.g., HLA) and pharmacogenes (e.g., CYP2D6).
HiFi target enrichment enables you to sequence at scale with ready-made Twist Alliance Panels or custom designs. Learn more about Twist and plan your project with this app brief.
HiFi amplicon sequencing
Enjoy a simplified workflow with one primer pair and one amplicon pool, easy assay optimization, long-range phasing, and simplified data analysis. Target medically relevant, individual genes or small gene panels using a PCR-based approach. If you can amplify it, HiFi sequencing can span your amplicon in a single read, giving unambiguous haplotype resolution through direct phasing. HiFi amplicon sequencing allows you to replace multiple assays with one fast, easy, scalable, and economical workflow.
Plan your project with this resource and learn more about our barcoding products.
Targeted sequencing datasets
Application note
HiFi targeted sequencing and comprehensive analysis of SMN1/2 with Paraphase
In this Application Note, we demonstrate targeted HiFi sequencing assays combined with a new informatics method for the combined analysis of SMN1 and SMN2. The software tool, Paraphase, is an informatics method that identifies full-length SMN1 and SMN2 haplotypes, determines gene copy numbers, and calls phased variants using PacBio® HiFi data.
Targeted sequencing in action
Video
Advancing pharmacogenomics research
HiFi sequencing enables comprehensive interrogation of pharmacogenomics genes such as single-gene assays or large panels.


NEW PRODUCT: HIFI TARGET ENRICHMENT WITH TWIST PROBES
Learn how you can get all the benefits of long and accurate HiFi reads at scale.
Long reads elucidate HIV envelope variants
Caskey and coworkers use targeted SMRT sequencing to determine that pre-existing variants in the HIV envelope may confer resistance to…
Identifying key players in host-microbiome interactions
HiFi sequencing of extended 16S amplicons has enabled the identification of metagenome community members at higher taxonomic…
EXPLORE
Did you know we have a comprehensive library of articles, reports, papers, and videos related to targeted sequencing?
Application brief
Scalable HiFi sequencing with Twist Bioscience long-read alliance panels
Access many benefits of PacBio® HiFi long-read sequencing at a fraction of the cost with Twist Bioscience long-read sequencing panels. Twist Bioscience’s target enrichment technology provides high capture efficiency and exceptional uniformity across target regions.
Resources
Application notes
- HiFi amplicon sequencing for congenital adrenal hyperplasia
- HiFi amplicon sequencing for Thalassemia
- HiFi amplicon sequencing for pharmacogenetics: CYP2D6
- HiFi targeted sequencing and comprehensive analysis of SMN1/2 with Paraphase
Application briefs
- HiFi target enrichment with Twist
- Targeted sequencing for amplicons
- Twist dark regions application brief
Protocols
Blog
Shining a light on dark genes
Countries around the world have recognized the value of precision health and population genomics research initiatives. They have launched efforts to sequence thousands to millions of genomes in order to learn more about how complex or rare inherited diseases impact their populations and how to mitigate their impacts. The goal? Building diverse health databases, learn how biology and lifestyle affect health, and bring us closer to a future where precision medicine is the standard.
But a population genomics program database can only be as good as the data it collects. Which raises the question – are the programs in place today able to collect everything they need to be the most effective for their target population?
Targeted sequencing workflow at a glance
Easy-to-use, high-throughput sequencing for the accuracy you need at an affordable cost
Straightforward library prep
Go from sample to sequencer in a single afternoon
Complete + accurate sequencing
Obtain accurate, unbiased, and long reads
Directly detect epigenetic modifications
Universal file types
Analyze data using any of the common genome assembly and analysis tools
YOUR MOMENT IS WAITING
Long reads at a lower cost and higher throughput. Short reads with near-perfect accuracy. From the fullest picture to the finest detail, your moment of discovery is waiting.