
Much has changed since the first insect genome assembly (Drosophila melanogaster) was published in 2000. With PAGBio Day 2022 registration kicking off this week, we thought it was the perfect time to take a look at how far we’ve come.
By November 2020, 601 different insect species representing 20 orders and sizes from the tiny 99 Mb Belgica antarctica to the massive 6.5 Gb Locusta migratoria, had nuclear genome assemblies available in GenBank.
And now, a new species of insect has its genome assembly deposited in GenBank every 2-3 days. Nearly 50% (n = 292) of the best-available insect assemblies were accessioned in 2019–2020. These new assemblies are markedly more contiguous than those of just a few years ago, due in part to the emergence of long-read assemblies, which rose in frequency from 0% of all assemblies in 2011–2012 to 36.1% in 2019–2020.
As Washington State University postdoc Scott Hotaling (@MtnScience) notes in his review of 20 years of insect, “we have entered a new era of insect genome biology.”
This is due largely in part to rapidly developing sequencing and analytical technologies that have brought the power of genome sequencing to an ever-expanding pool of researchers, he said.
“Most notable has been the impact of long-read sequencing; assemblies that incorporate long reads are ∼48× more contiguous than those that do not.”
The addition of low and ultra-low DNA input workflows on Sequel II Systems has benefited entomologists even further by facilitating the sequencing of even the smallest species. Just 5 ng of genomic DNA is all that is needed, enabling the creation of high-quality genomes from single bugs, without the need for time-consuming inbreeding or pooling strategies.
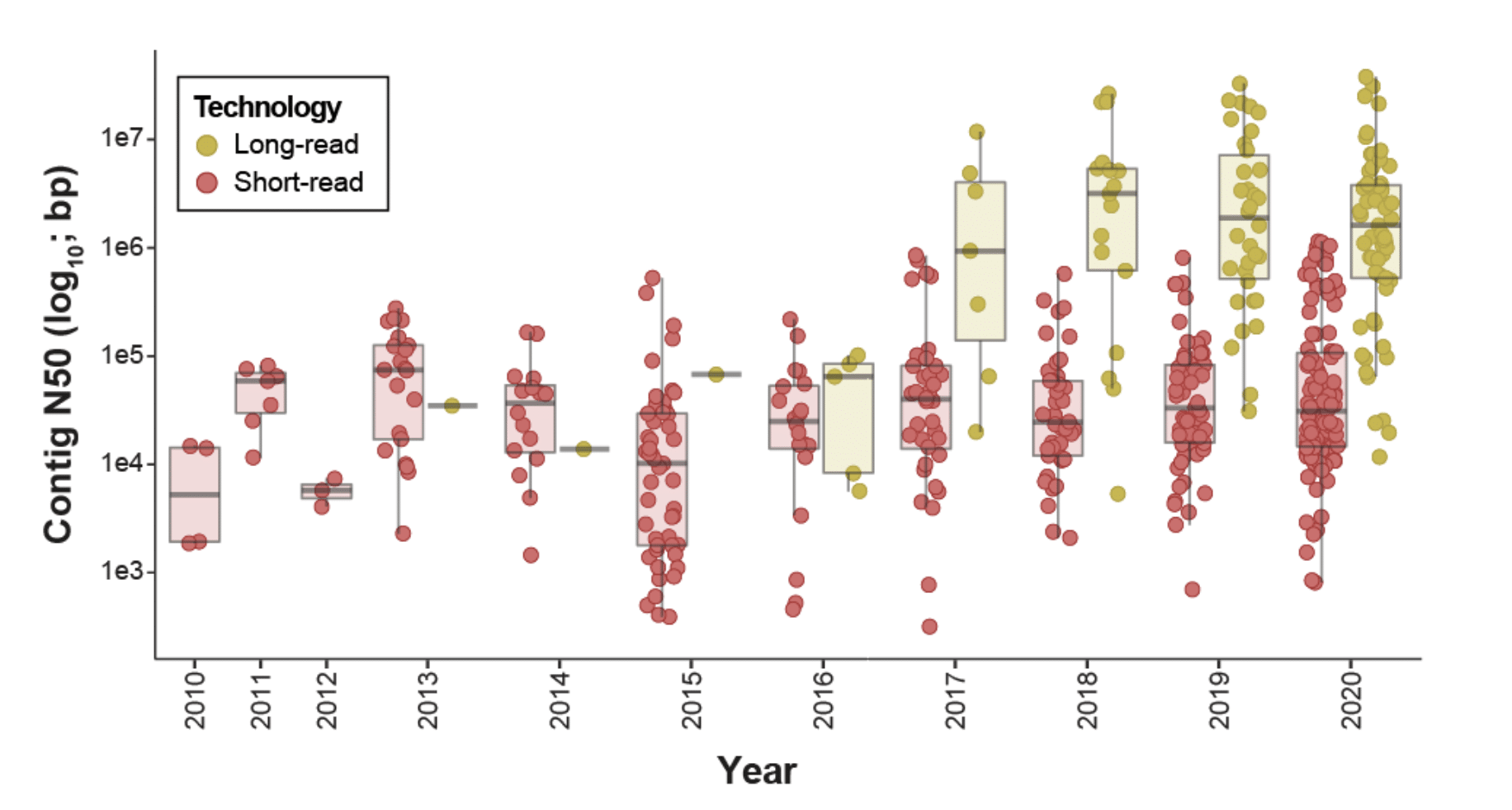
Go bigger
Published in the journal Genome Biology and Evolution with co-author Paul B Frandsen (@paulbfrandsen) of Brigham Young University, LOEWE Centre for Translational Biodiversity Genomics (LOEWE‐TBG) and the Smithsonian Institute, the review also includes a call out to peers to make the insect genomic record even more complete.
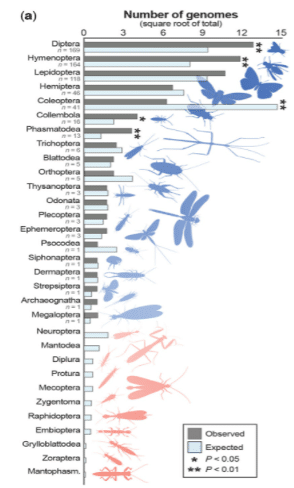
Although 600+ taxa may seem impressive, there are many taxonomic gaps, the researchers note.
“Aquatic insects, as a group, are underrepresented relative to their terrestrial counterparts. And, some orders (e.g., Diptera) are represented by far more genome assemblies than their species diversity alone would warrant—likely reflecting the model organisms within them—although many orders still have no genomic representation,” they write.
By the numbers:
● Relative to species richness, genomic efforts have been biased toward four orders: Diptera, Hymenoptera, Collembola, and Phasmatodea.
● Coleoptera, with 387,100 described species, are significantly underrepresented (41 assemblies vs. ∼228 expected).
● Six orders were represented by only one genome assembly and 11 orders had no publicly available assembly.
The authors call for increased integration between independent research groups and consortia like the i5K initiative to sequence genomes for 5,000 different arthropods, as well as strategic sampling to fill taxonomic gaps and generate data for targeted questions, and expanded gene annotations.
As of 2019, only 40% of insect genome assemblies had corresponding gene annotations in GenBank; expanding and refining the availability of gene annotations for insects will drive corresponding increases in the scale of taxonomic comparisons that are possible for many analyses, the authors argue.
HiFi sequencing allows for quick assembly of complex organisms and can help scientists establish more diverse datasets of clades.
“With genome assemblies representing 600+ taxa and ∼480 Myr of evolution available in a public repository, the power and promise of insect genome research has never been greater,” Hotaling concluded.
“We echo the findings of the Vertebrate Genome Project—long-read assemblies are vastly more contiguous than short-read approaches—and recommend that these technologies be embraced by insect genome scientists.”
Stay tuned, the field of insect genomics is rapidly expanding and HiFi reads play a key role in establishing genomic data that represents the true diversity of the arthropod world.
Read more about the low & ultra-low DNA input workflows:
Don’t Sweat The Small Stuff: Low DNA Input Workflow Enables Sequencing of the Smallest Species
New Low-Input Protocol Enables High-Quality Genome Created from Single Mosquito
Sequencing at the Extremes: Low DNA Input Workflow Enables Study of Tiny Ice Worm with Giant Genome
Procedure & Checklist: Preparing HiFi Libraries from Low DNA Input Using SMRTbell Express Template Prep Kit 2.0
Now Available: Ultra-Low DNA Input Workflow for SMRT Sequencing
Application Note: Considerations for Using the Low and Ultra-Low DNA Input Workflows for Whole Genome Sequencing