Recent de novo assemblies of individual human genomes have uncovered thousands of structural variants, many of which are accessible only with PacBio long reads [1-3].
| Personal Genome | PacBio Coverage | Deletions ≥50 bp | Insertions ≥50 bp |
| CHM1 [1] | 41-fold | 6,111 | 9,638 |
| HX1 [2] | 103-fold | 9,891 | 10,284 |
| AK1 [3] | 101-fold | 7,358 | 10,077 |
A similar increase in structural variant sensitivity relative to short-read methods has been demonstrated with low-fold coverage PacBio sequencing interpreted against the reference genome [4]. To demonstrate and evaluate the low-fold coverage approach on the PacBio Sequel System, we generated approximately 10-fold coverage of the well-studied human sample NA12878.
Methods
Purified DNA for NA12878 was obtained from Coriell, sheared to an average size of 25 kb, converted to SMRTbell templates, and size selected to 15 kb on the BluePippin system (Sage Science). The resulting library was loaded on 10 SMRT Cells. Each SMRT Cell was run for 6 hours on the Sequel System with chemistry v1.2 (an older chemistry than was used for recently released Arabidopsis data, which uses the newer chemistry v1.2.1 and has a yield of about 5 Gb per SMRT Cell and read length N50 of 16.4kb). In total, the runs generated 32.8 Gb of data contained in 3.4 million reads with half of the bases in reads longer than 11.8 kb.
Sequencing Metrics
| SMRT Cells | 10 |
| Run Time | 60 hrs |
| Number of Bases | 32.8 Gb |
| Number of Reads | 3.4 M |
| Read Length N50 | 11,823 bp |
Reads were mapped to the GRCh37 human reference genome with NGM-LR [5], and structural variants were called with PBHoney [6]. A total of 7,386 deletions and 7,445 insertions of at least 50 bp were identified and comprise the “10-fold SV call set.”
Visualizing Structural Variants
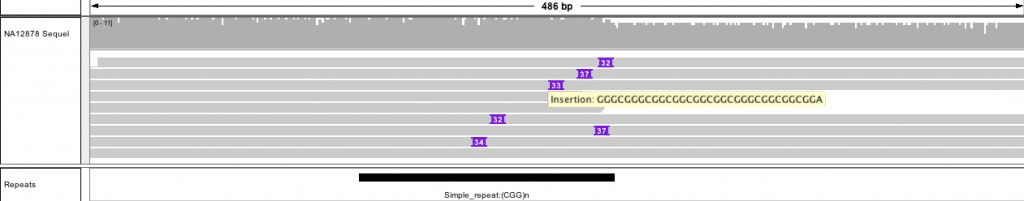
Ongoing improvements to the IGV browser [7] (available now in the development version) improve visualization for PacBio reads and structural variants. With these updates, IGV provides a clear representation of deletions, insertions, and trinucleotide repeats, and shows how long reads span structural variants.
Heterozygous 315 bp deletion at chrX:116,454,160-116,454,859
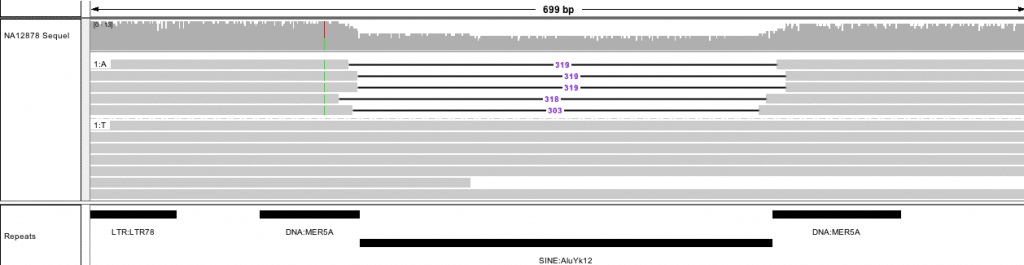
Homozygous 328 bp insertion at chr10:92,213,800-92,216,245

FMR1 trinucleotide repeat small expansion at chrX:146,993,200-146,993,950
Evaluation of 10-fold Call Set
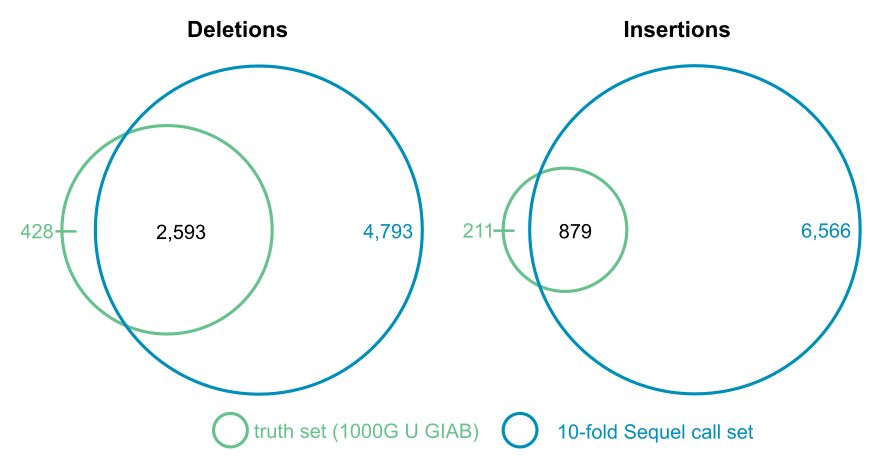
To quantify sensitivity, the 10-fold SV call set was compared to a merged NA12878 “truth” set from the 1000 Genomes Project [8] and Genome in a Bottle [9].
| Set | Platform | Deletions ≥50 bp | Insertions ≥50 bp |
| truth: 1000 Genomes + GIAB [8,9] | Illumina | 3,021 | 1,090 |
| 10-fold SV call set | PacBio Sequel | 7,386 | 7,445 |
The 10-fold SV call set recalls 86% of truth set deletions and 81% of insertions. Moreover, it includes thousands of deletions and insertions that are not in the truth sets, most of which are directly confirmed by a FALCON-Unzip de novo assembly from 60-fold PacBio RS II coverage.
In summary, this 10-fold SV call set demonstrates that low-fold coverage sequencing on the PacBio Sequel System is an affordable, effective approach for identifying structural variants and provides much improved sensitivity compared to short-read approaches. We are excited to see how this approach will be extended and applied to study genetic variation in disease cohorts, in human populations, and in other organisms.
Data Availability
To illustrate the low-fold coverage structural variant calling workflow, the NA12878 Sequel data is available for analysis on DNAnexus.
[1] Chaisson MJ, et al. (2015). Nature, 517(7536):608-11.
[2] Shi L, et al. (2016). Nat Commun, 7:12065.
[3] Seo JS, et al. (2016). Nature, 538(7624):243-7.
[4] English AC, et al. (2014) BMC Bioinformatics, 15:180.
[5] https://github.com/philres/nextgenmap-lr
[6] English AC, et al. (2015). BMC Genomics, 16:286.
[7] Robinson JT, et al. (2011). Nat Biotechnol, 29(1):24-6.
[8] Parikh H, et al. (2016). BMC Genomics, 17:64.
[9] Sudmant PH, et al. (2015). Nature, 526(7571):75-81.