Confident and complete resolution of viral diversity
HiFi sequencing provides 99.9% accurate reads from amplicons up to 20 kb, enabling direct detection of phased variants and giving you the ability to:
- Deconvolute complex mixtures into quasispecies and unique haplotypes
- Track the evolution and phylogeny of viral populations in a host, within a community, or across geographic regions
- Identify and quantify minor variants linked to immune evasion or drug resistance
- Generate complete de novo assemblies of large viral genomes
Workflow: from DNA to resolved viral populations
Sample + library preparation
Obtain targeted sequences with flexible sample and library workflows.
- Library input as low as 250 ng of viral genomic DNA or cDNA
- Sequence amplicons up to 20 kb with 99.9% accuracy with HiFi reads
- Scale throughput as needed with our barcoded adapter kits or M13 primer plates
Sequencing
Scale coverage based on your project needs with the Revio and Sequel II systems
- Achieve 2 – 4 million or 5 – 10 million HiFi reads per SMRT Cell 8M on a Sequel II system or per SMRT Cell 25M on a Revio system, respectively
- Maximize output and turn-around-time with adjustable run parameters
- Choose 10 – 30 hr movie times based on amplicon or insert size
*Read lengths, number of reads, data per SMRT Cell, and other sequencing performance results vary based on sample quality/type and insert size, among other factors.
Data analysis
Make discoveries using bioinformatics tools in SMRT Analysis or PacBio DevNet.
Read our SMRT Analysis Barcoding Overview
- To phase SNPs and determine the frequency of minor variants in a viral population, use juliet, our minor variant tool
- To resolve a population of pooled amplicons, use our Long Amplicon Analysis pipeline
Spotlight
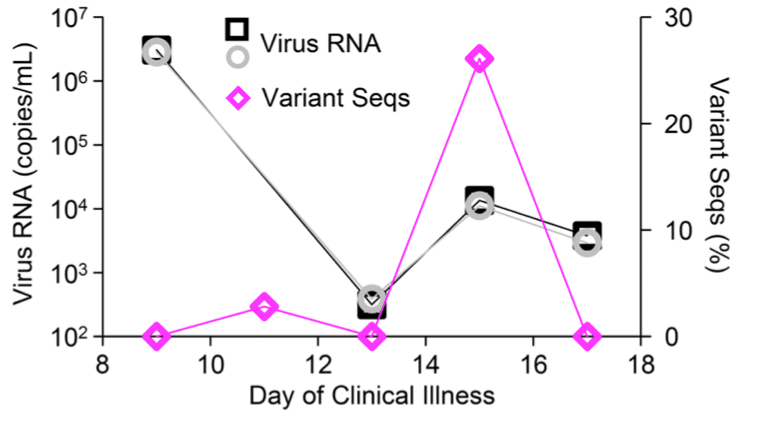
HiFi reads reveal emergence of SARS-CoV-2 minor variants during acute infection
A longitudinal study of a person who ultimately recovered showed that spike variants resistant to autologous neutralizing antibodies arose transiently before being displaced by the original strain ahead of virus clearance. Application of the method to more diverse cohorts may reveal circumstances under which variants capable of immune escape emerge and persist.
Watch Dr. Eli Boritz present his research at our webinar, Beyond COVID-19: The Long and the Short of Why Virologists Use HiFi Sequencing.
Spotlight
ADVANCING VACCINE RESEARCH BY LINKING QUASISPECIES GENOTYPE TO PHENOTYPE
PacBio single-molecule sequencing was used to characterize how an attenuated candidate RSV strain, Min B, evolved with serial passage under selection pressure. Targeted sequencing revealed the rapid emergence of defective ‘helper genomes’ with large deletions (LD RNAs) that enabled the population as a whole to regain replication competence.
Nouën C.L., et. al. (2021) Rescue of codon-pair deoptimized respiratory syncytial virus by the emergence of genomes with very large internal deletions that complemented replication. PNAS
Infographic
Mapping the interplay of viral and host genomics with HiFi sequencing
Learn how you can use HiFi reads to enable your viral research, from understanding viral genomes to the host immune response.