Flexible workflows for all your important genomic targets
Single Molecule, Real-Time (SMRT) sequencing offers a flexible solution for targeting regions of interest and delivers the most comprehensive view of the associated genes using highly accurate long reads (HiFi reads). SMRT sequencing gives you the ability to:
- Rapidly screen and identify all variants
- Discover haplotype-specific markers
- Resolve difficult-to-sequence regions, regardless of size
- Confirm insertion sites of transgenes and validate gene editing events
Interested in using an amplification-free approach? Explore No-amp targeted sequencing to enrich and sequence hard-to-amplify genomic regions.
Workflow: from sample to base-level resolution
Sample + library preparation
Obtain targeted sequences with flexible sample and library workflows
Easy-to-use library preparations and multiplexing for all experimental designs:
- Start with a minimum of 250 ng of high-quality nucleic acids
- Create SMRTbell templates from amplicons up to 20 kb
- Optimize throughput with flexible barcoding options and multiplexing up to 10,000 samples per SMRT Cell
- Amplify PCR products using target-specific primers with incorporated barcodes
- Add barcoded universal primers into amplicons via a simple 2-step PCR process
- Attach barcoded overhang adapters during ligation without modifying existing primers
Sequencing
Maximize output and turn-around time with adjustable run parameters on the Sequel systems.
Sequence to desired coverage based on project needs
- Maximize output and turn-around-time with adjustable run parameters
- For inserts <5 kb, recommend 10-hour movies
- For inserts >5 kb, recommend 20-hour movies
- Generate up to 500,000 Q20 HiFi reads per SMRT Cell 1M
- Sequence to desired coverage based on project needs
- Target 30-fold coverage for variant detection
*Read lengths, number of reads, data per SMRT Cell, and other sequencing performance results vary based on sample quality/type and insert size, among other factors.
Data analysis
Make discoveries using bioinformatics tools with the PacBio analytical portfolio.
- Fully characterize genetic complexity – structural variation, rare SNPs, indels, CNV, microsatellites, haplotypes, and phasing
- Utilize a variety of analysis tools within SMRT Link with outputs in standard file formats (BAM and FASTA/Q)
- Generate HiFi reads (>Q20) using circular consensus sequencing (CCS) analysis
- Perform reference-free analysis of pooled amplicons with long amplicon analysis (LAA)
- De-multiplex up to 384 barcodes; command line supports >384 barcodes
- HiFi reads are compatible with standard analysis tools for variant calling such as GATK
Application brief
Targeted sequencing for amplicons best practices
Learn more about these best practices for targeted sequencing of amplicons.
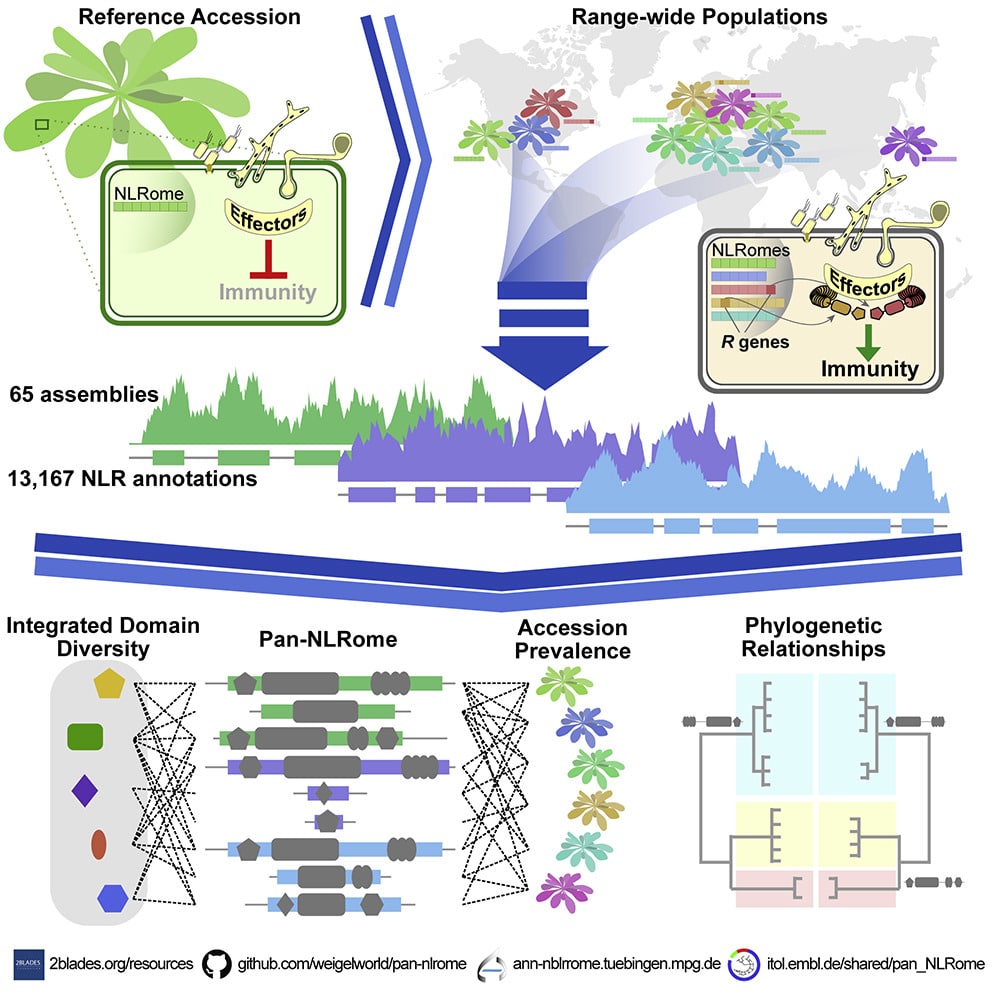
Mapping the NLRome
Research teams turn to SMRT sequencing to trace plant immunity
In a recent paper, a multi-institutional team detailed their creation of a nearly complete species-wide database of nucleotide-binding leucine-rich repeat receptors, or NLRs, in Arabidopsis thaliana. The sequences they obtained allowed them to define the core NLR complement, as well as to chart integrated domain diversity, describe new domain architectures, assess presence or absence of polymorphisms in non-core NLRs, and map uncharacterized NLRs onto the A. thaliana Col-0 reference genome. Explore this research further:
Van de Weyer, A.-L., et al. (2019). A Species-Wide Inventory of NLR Genes and Alleles in Arabidopsis thaliana. Cell, 178(5), 1260–1272.e14.
Spotlight
SMRT Sequencing Reveals Biodiversity
“The Sequel II represents a key component in the support infrastructure for BIOSCAN, a new seven-year $180 million research program which will advance our understanding of biodiversity by accelerating species discovery, and by revealing species interactions and dynamics. Given our plans to barcode tens of millions of specimens, it’s critical that analytical costs be reduced and the Sequel II makes this possible while also delivering very high fidelity sequences.”