Long-read sequencing myths: debunked.
Part 3 — cancer genomics

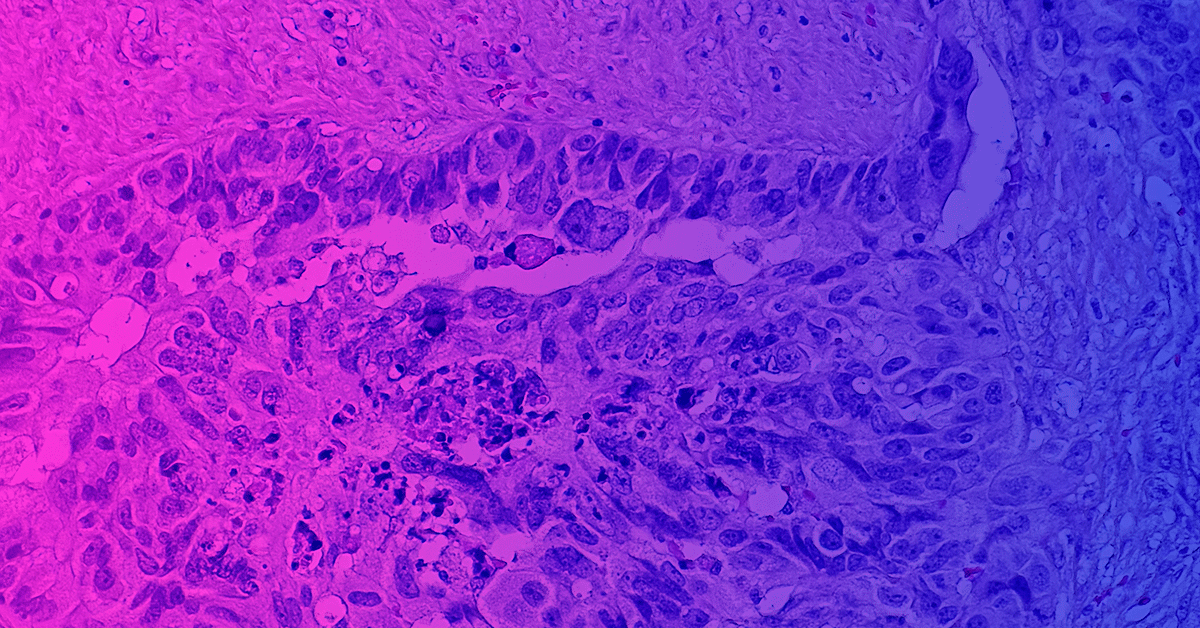
The complexities of cancer biology are famously tricky to decipher. Peering into the murky cancer genome and making sense of tumor mechanisms has long been the goal of researchers…