In order to help evaluate the utility of long, unbiased sequence reads for characterizing structural variation in the human genome using our recently released P5-C3 scaffolding sequencing chemistry, we have collected 10x long-read, shotgun coverage of a human genome sample. The human genome harbors many structural variations, including variable number tandem repeats, deletions, insertions, inversions, and repetitive mobile elements, which are often difficult to resolve using short-read technologies. We hope this data set will be of value to the bioinformatic and scientific community studying various forms of structural variation across the human genome. To access the full data set, simply send us an email and you will receive instructions for downloading.
In collaboration with Evan Eichler (Howard Hughes Medical Institute, University of Washington), we sequenced CHM1TERT, a well-studied cell line derived from a complete hydatidiform mole (CHM). A hydatidiform mole is defined as a pregnancy with no embryo and clinically presents in approximately 1 in 1,500 pregnant women in North America. The CHM cells have a diploid genome, typically XX, that is a result of replication of a haploid paternal (sperm) genome. Through the corresponding absence of allelic variation, this sample has been used to generate a haploid reference genome sequence, and many associated resources are available, including physical maps, genotypes (iSCAN), and a large-insert BAC library (CHORI-17). It is also one of the targets for the production of a higher quality “platinum” genome assembly.
We prepared ~20 kb DNA fragment libraries, size-selected with the BluePippin™ system from Sage Science, and sequenced with 3-hour movies using the P5-C3 sequencing chemistry. Some sequencing statistics are listed below:
- Total number of reads: 3,679,463
- Total number of post-filtered bases: 32,559,803,198
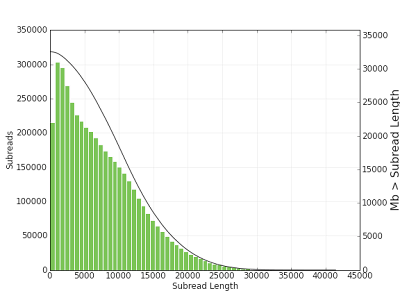
- Average read length: 8,849 bp
- Half of sequenced bases in reads greater than: 10,985 bp
- 5% of sequenced DNA inserts longer than: 18,060 bp
- Longest DNA insert sequenced: 41,460 bp
- PacBio® RS II instrument time for sequencing: 10 days
- Number of SMRT® Cells: 66
Figure 1. Subread length distribution. A subread is a DNA insert sequenced between two SMRTbell™ hairpin adapters. The solid black line (right y axis) denotes the amount of sequenced bases greater than a given subread length (x axis).
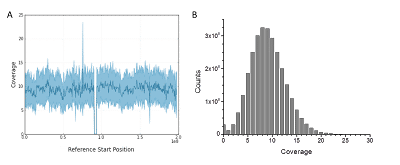
We also mapped the data against the human reference genome (GRCh37) and found generally even coverage across the reference, with numerous examples of structural variations highlighted by the long reads. A mapping coverage summary and a few examples highlighting structural variation are given below.
Figure 2. Uniform sequencing coverage upon mapping against the GRCh37 human genome reference. (A) Example coverage for chromosome 3. The gap in the center is due to lack of sequence in the reference (~3 million N bases) of the centromere. (B) Coverage histogram over all non-N bases of the GRCh37 reference.
Figure 3. Examples of large deletions. The sharp breakpoints from the even shotgun read structure, combined with the lack of read coverage, indicate a 114.2 kb and a 4.9 kb deletion in this ~375 kb region of chromosome 3. The individual sequence reads are shaded by length (reads in black are >10 kb). Both deletions have been validated and are polymorphic in the human population.
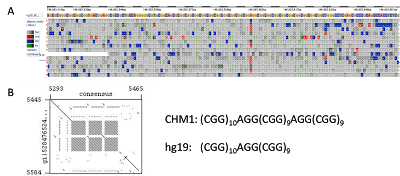
Figure 4. Sequence structure of the Fragile X Mental Retardation (FMR1) Triplet CGG Repeat. (A) Read mapping to the reference genome sequence shows many insertions (green vertical lines) across this region on the X chromosome. (B) Consensus building from the reads and dot plot comparison reveals the true structure including an additional AGG-(CGG)9 repeat block in the CHM1 genome.