Accurate variant detection even in complex regions of the genome
Single Molecule, Real-Time (SMRT) sequencing enables you to detect all variant types such as structural variants, rare SNPs, indels, copy number variation, microsatellites, haplotypes, and phased alleles while sequencing through low complexity regions like repeat expansions, promoters, and flanking regions of transposable elements. Explore the targeted sequencing applications and workflows.
Targeted sequencing
Utilize highly accurate long reads (HiFi reads, >99% single-molecule read accuracy) to sequence targets ranging in size from several hundred base pairs to 20 kb. Choose between push-button solutions in SMRT Analysis or flexible command-line tools utilizing industry standard workflows.
SMRT Analysis
Our three targeted sequencing analysis applications allow you to explore structural variants, rare SNPs, indels, copy number variation, microsatellites, haplotypes, and phased alleles:
- Circular consensus sequencing
- Long amplicon analysis
- Minor variant analysis
Learn more about SMRT Analysis and structural variant calling
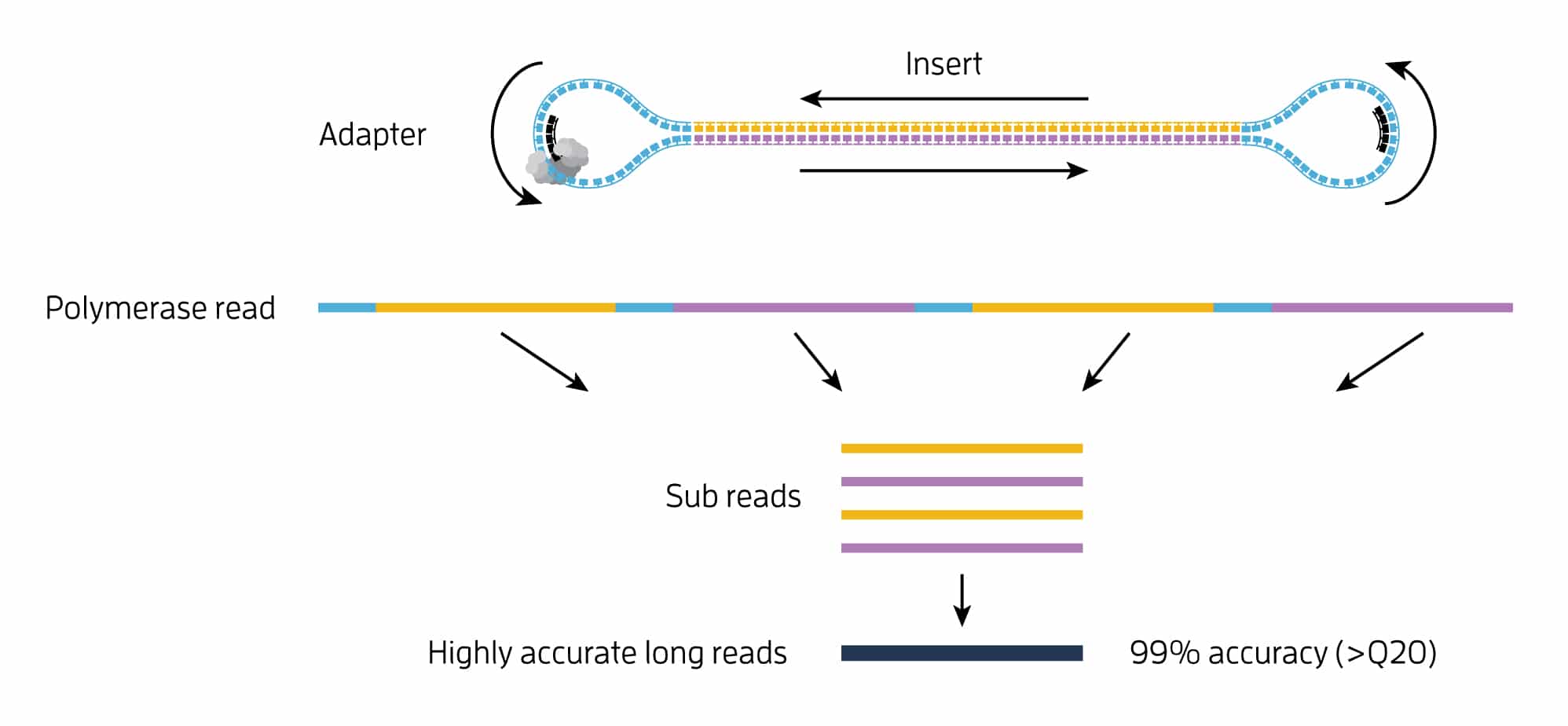
Circular consensus sequencing (CCS)
The CCS algorithm generates HiFi reads with >99% accuracy and length up to 20 kb. Resulting HiFi reads can be used for variant calling and phasing in SMRT Analysis or other tools such as GATK, DeepVariant, and WhatsHap.
How it works: CCS generates a highly accurate reads based on building consensus from the multiple passes of forward and reverse strands of the insert DNA incorporated into a SMRTbell molecule.
PB Amplicon Analysis (pbAA)
Generates phased consensus sequences by clustering HiFi reads. The output of pbAA is ideal for diplotyping complex loci of biomedical interest such as HLA genes or CYP2D6.
How it works:
HiFi reads from sequenced amplicons are separated by locus and allele using customized “guide” sequences supplied by the user. Reads are clustered and a consensus sequence is produced for each cluster. Consensus sequences that pass quality filters can be used with downstream workflows such as mapping, variant calling, and allele assignment.
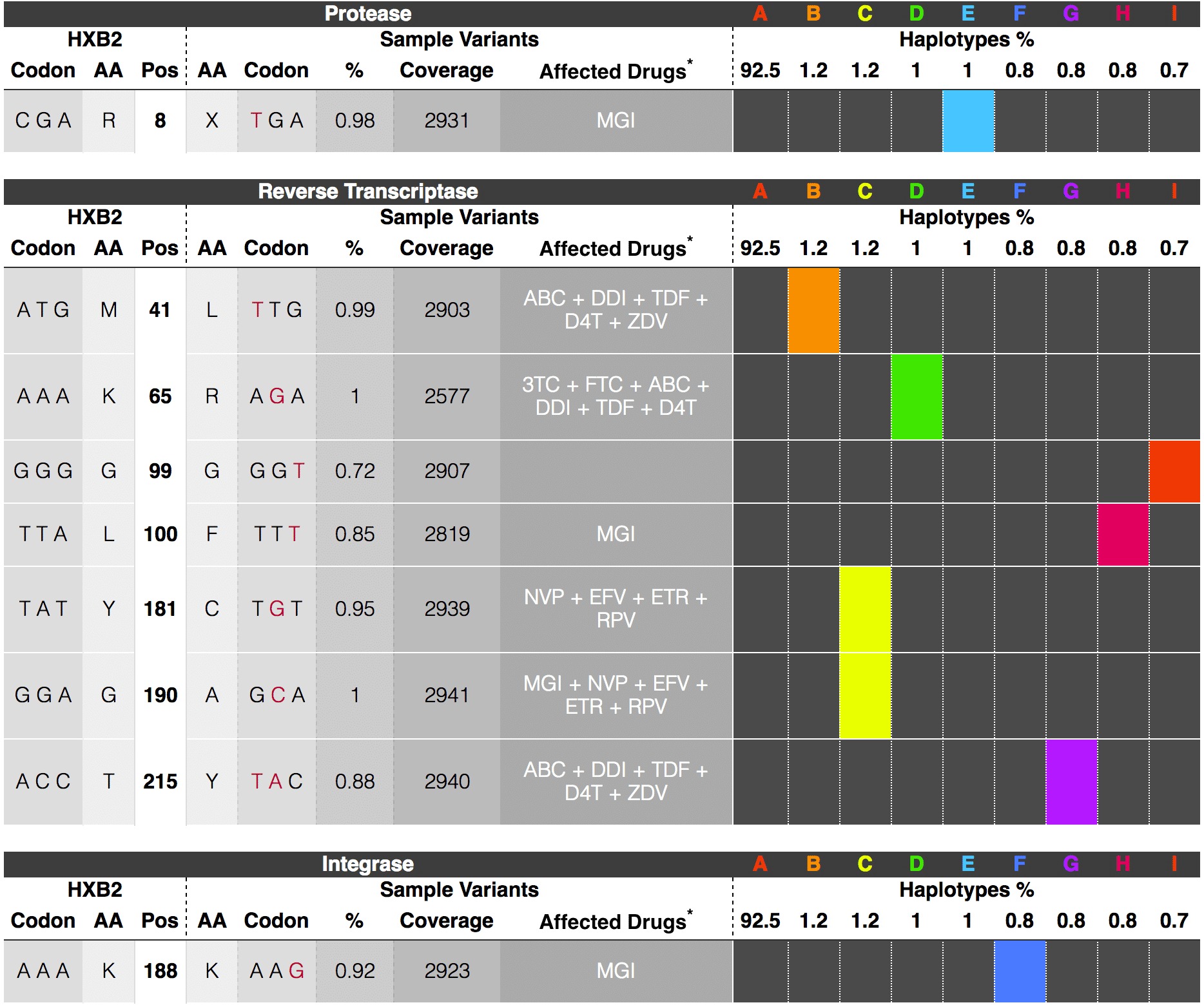
Minor variant analysis
This application robustly identifies and phases minor single nucleotide substitution variants, with frequencies as low as 1%, from complex populations with false positive and negative rates below 1% and 0.001%, respectively. The general use reference-guided de novo variant caller phases codon variants into haplotypes. It calls and annotates variants in ORFs but does not support small indels.
How it works: HiFi reads generated with CCS are aligned to a reference sequence, amino acid changes are identified using statistical test. Drug resistance can be identified if annotation reference is available.
PacBio DevNet tools
Additional options for data analysis include community-supported tools for alignment, variant detection, phasing, etc. They provide scientists with easy access to open-source algorithms such as:
GATK
A toolkit developed by the Broad Institute, that offers a wide variety of tools with a primary focus on variant discovery and genotyping
DeepVariant
An analysis pipeline developed by Google that uses a deep neural network to call genetic variants from next-generation DNA sequencing data
See our complete list of PacBio DevNet tools for targeted sequencing
SMRT compatible analysis products:
Our analysis partners offer several solutions for analysis of targeted sequencing data. Options include software applications, full-service genomics analytics, or cloud-based platforms with open source tools.
Golden Helix
SNP & Variation Suite is an integrated collection of user-friendly and powerful analytic tools for managing, analyzing, and visualizing multifaceted genomic and phenotypic data
Partek Genomics suite
A user-friendly, start-to-finish software solution designed for the integration and analysis of all genomic data
See our complete list of SMRT compatible analysis products
