Resolve the full spectrum of genetic variation
Single Molecule, Real-Time (SMRT) sequencing delivers a complete, accurate view of the clonal distribution of mutations in genes or genomic regions of interest. The PacBio systems produce reads long enough to span full-length transcripts and to allow for the immediate detection of compound mutations and splice isoforms. With these sequencing technologies, you have the ability to:
- Detect SNVs occurring at a frequency as low as 1%
- Differentiate polyclonal from compound mutations
- View splice variants in gene fusion transcripts
- Discover every breakpoint in regions of genomic instability
Workflow
From sample to somatic variant detection
Library preparation
- Universal SMRTbell template for insert sizes from 2 kb to 10 kb
- Size-selection options enrich for longest inserts
- Library automation supported
- Additional target capture and enrichment strategies available
- Multiplex 384 barcoded samples
SMRT sequencing with PacBio systems
- Take advantage of the Sequel system to reduce project costs and generate 7× more reads compared with the PacBio RS II
- Achieve ~10 kb average read lengths, with some reads as long as 60 kb
- Adjust run times (0.5 to 6 hours) to maximize sample throughput and turn-around time
- Obtain consensus accuracies >99.999% by avoiding mapping and systematic errors
- Produce high, single-molecular consensus accuracies through multiple observations of single circularized templates
Data analysis with SMRT Analysis and PacBio DevNet
- Long amplicon analysis (LAA) for generation of reference-free de novo sequences from pooled amplicons
- Minor variant analysis for detection and quantification of single nucleotide polymorphisms
- Iso-Seq for characterization of transcripts and isoforms
- ClusterConsensus for reference-free deconvolution of genomes in a complex mixture
- HGAP for comprehensive de novo assembly of BACs or fosmids
Spotlight
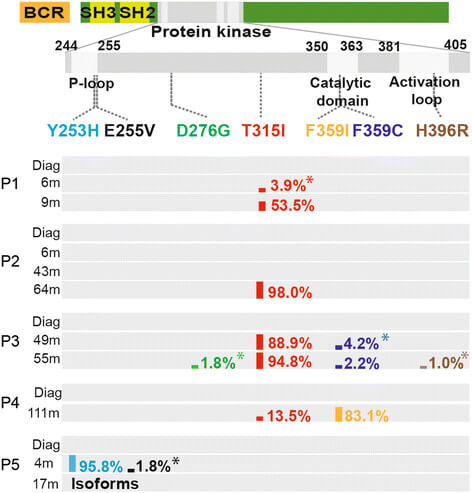
A comprehensive view of BCR-ABL1 fusion gene mutations
SMRT sequencing provided a complete view of the clonal distribution of mutations in the BCR-ABL1 fusion gene implicated in chronic myeloid leukemia treatment resistance. The authors identified several mutations and novel transcript isoforms standard clinical assays had missed. PacBio sequencing provided clonal distribution frequencies for the compound mutations and isoforms. Explore this research further:
Cavelier, L. et al. (2015) Clonal distribution of BCR-ABL1 mutations and splice isoforms by single-molecule long-read RNA sequencing. BMC Cancer. 15, 45.